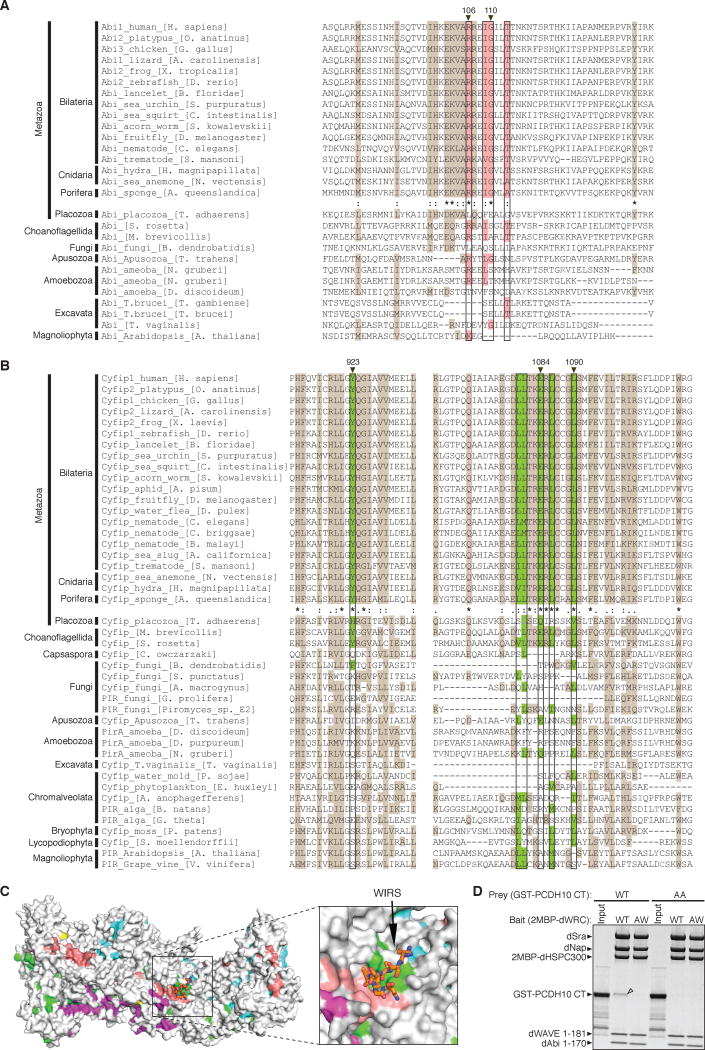
Figure 3. WIRS Binding Surface is Highly Conserved.
(A–B) Sequence alignments of representative organisms. Surface residues of the WIRS binding site (black boxes) are highlighted with pink for Abi (A) and green for Sra1 (B). Other conserved residues were highlighted with brown. Degrees of conservation up to Porifera are represented with ClustalW symbols (* for no change, : for conserved, . for less conserved changes). Residues whose mutation disrupts WIRS binding in Figure 2 are labeled with black triangles on top. Grey amino acids indicate where sequence insertions in alignment were not shown.
(C) Surface conservation of the WRC, with the most conserved surface residues (ConSurf score 9) (Ashkenazy et al., 2010) colored as in Figure 2A and less conserved residues (ConSurf score < 9) in grey.
(D) Commassie blue stained SDS-PAGE gel shows that immobilized wild type Drosophila WRC (WT), but not a mutant with a disrupted WIRS binding surface (AW for R118A/G122W-Abi) selectively retained GST-PCDH10 CT (WT), but not a mutant (AA for T1002A/F1003A).