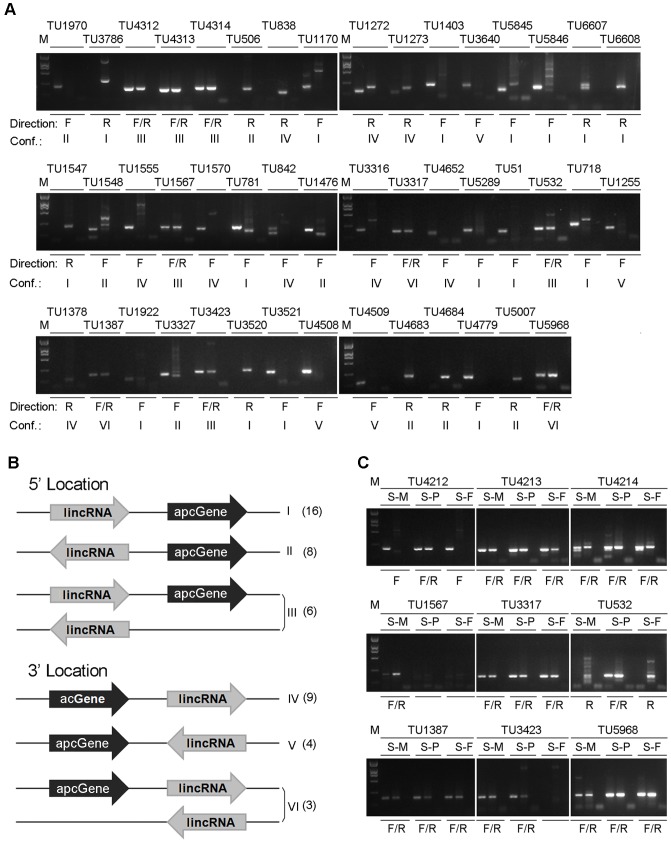
Figure 3. Determination of transcript orientation for 46 lincRNAs by MRA.
(A) Electrophoresis analysis of the products amplified from pooled RNA samples. The products from the second-round PCR were separated by electrophoresis with 2% agarose gels in the order of P-4a, P-4b, and P-c as described in Figure 2-(5). The IDs of lincRNAs are listed above the gel picture, and the transcriptional direction and lincRNA/apcGene configuration are listed below the gel picture. The explanation of each configuration is shown in panel (B) M: DNA marker DL2000; F: forward; R: reverse; F/R: bidirectional; Conf: lincRNA/apcGene configuration; (B) Schematic representation of the various types of lincRNA/apcGene configurations. The lincRNA is shown as the gray arrow, whereas the apcGene is shown as the black arrow. The IDs for the configurations are represented by roman numerals. The number of lincRNAs belonging to each configuration is shown in parenthesis; (C) Gel electrophoresis of the nine bidirectional lincRNAs using RNA samples isolated from three developmental stages separately. S-M: RNA sample from mycelia; S-P: RNA samples from primordia; S-F: RNA sample from fruiting bodies.