Abstract
CSPG4 marks pericytes, undifferentiated precursors and tumor cells. We assessed whether the shed ectodomain of CSPG4 (sCSPG4) might circulate and reflect potential changes in CSPG4 tissue expression (pCSPG4) due to desmoplastic and malignant aberrations occurring in pancreatic tumors. Serum sCSPG4 was measured using ELISA in test (n = 83) and validation (n = 221) cohorts comprising donors (n = 11+26) and patients with chronic pancreatitis (n = 11+20) or neoplasms: benign (serous cystadenoma SCA, n = 13+20), premalignant (intraductal dysplastic IPMNs, n = 9+55), and malignant (IPMN-associated invasive carcinomas, n = 4+14; ductal adenocarcinomas, n = 35+86). Pancreatic pCSPG4 expression was evaluated using qRT-PCR (n = 139), western blot analysis and immunohistochemistry. sCSPG4 was found in circulation, but its level was significantly lower in pancreatic patients than in donors. Selective maintenance was observed in advanced IPMNs and PDACs and showed a nodal association while lacking prognostic relevance. Pancreatic pCSPG4 expression was preserved or elevated, whereby neoplastic cells lacked pCSPG4 or tended to overexpress without shedding. Extreme pancreatic overexpression, membranous exposure and tissuehigh/seralow-discordance highlighted stroma-poor benign cystic neoplasm. SCA is known to display hypoxic markers and coincide with von-Hippel-Lindau and Peutz-Jeghers syndromes, in which pVHL and LBK1 mutations affect hypoxic signaling pathways. In vitro testing confined pCSPG4 overexpression to normal mesenchymal but not epithelial cells, and a third of tested carcinoma cell lines; however, only the latter showed pCSPG4-responsiveness to chronic hypoxia. siRNA-based knockdowns failed to reduce the malignant potential of either normoxic or hypoxic cells. Thus, overexpression of the newly established conditional hypoxic indicator, CSPG4, is apparently non-pathogenic in pancreatic malignancies but might mark distinct epithelial lineage and contribute to cell polarity disorders. Surficial retention on tumor cells renders CSPG4 an attractive therapeutic target. Systemic ‘drop and restoration’ alterations accompanying IPMN and PDAC progression indicate that the interference of pancreatic diseases with local and remote shedding/release of sCSPG4 into circulation deserves broad diagnostic exploration.
Introduction
Pancreatic cancer is one of the most aggressive therapy-resistant gastrointestinal malignancies. Because of the often late diagnosis, surgery represents a curative option for only 20% of patients, but this does not preclude recidivism or metastasis [1]. Novel diagnostic and therapeutic approaches are urgently needed. Pancreatic cancer is associated with a prominent desmoplastic reaction of prognostic and diagnostic relevance [2]–[4]. In a mouse model of PDAC, depletion of a rare population of the FAP+-fibroblasts/pericytes has been found to be sufficient to induce IFNg/TNF-mediated hypoxic (ischemic) necrosis of cancer cells and stroma via vascular damage and alleviated local immunosuppression [5]. Pericytes are mesenchymal cells seeding in the walls of newly formed blood vessels, playing a major role in angiogenesis, and representing one of the possible sources of the stroma-producing myofibroblasts in pancreatic cancer [6]. They are marked by chondroitin sulfate proteoglycan 4 (CSPG4/NG2), also known to decorate immature precursors of epidermal, neural, and mesenchymal origin [7]–[10].
Transmembrane CSPG4 functions as a high-affinity receptor for bFGF, PDGF-AA, alpha3beta1-integrin, galectin-3, and, most prominently, type VI collagen (COL6) [11]. The ligand-receptor interaction activates small GTPases; it can provide links to a number of cytoplasmic signaling pathways, enabling dynamic rearrangement of the actin cytoskeleton, altering cell-stratum interactions and promoting the process of migration. CSPG4 has also been found in certain tumor cells, and has been shown to promote their malignant behavior and to impact the progression of melanoma, glioblastoma, chondrosarcoma and leukemia [11]–[15]. Therapeutic targeting of CSPG4 in melanoma and glioblastoma appears to yield anti-tumor effects [16]–[18].
The extracellular portion of CSPG4 (ectodomain) may undergo different post-translational modifications [19], or be shed, assuming that autocrine proteolysis is not blocked by simultaneously produced COL6, which contains a Kunitz-type proteinase inhibitor sequence in the a3-chain [20]. The presence of ectodomain in circulation (sCSPG4), and its clinical relevance have not yet been evaluated. Although pro-stromal and pro-malignant, CSPG4 was not yet considered as a pathogenic factor or biomarker in pancreatic cancer; a stroma-rich aggressive malignancy [3], [21]. By studying the patients with non-neoplastic, benign and malignant pancreatic disorders, we sought to determine whether different degrees of malignant or desmoplastic transformation modify patterns of CSPG4 expression in tissues or circulation, and whether it might bear diagnostic or pathogenic relevance.
Materials and Methods
Serum and Tissue Sampling
The analyses included the pancreatic biopsies and sera from donors and patients with chronic pancreatitis or different variants of exocrine pancreatic tumors: benign, premalignant and malignant. The study was approved by the Ethics Committee of the Faculty of Medicine, University of Heidelberg, Germany (Vote 301/2001 and 159/2002) and performed with patients’ written informed consent and in compliance with institutional regulations. Freshly removed tissues were flash-frozen in liquid nitrogen for RNA and western blot profiling, or fixed in paraformaldehyde solution for 12–24 h prior to paraffin embedding for histological analysis. Serum sCSPG4 was measured using ELISA in test (n = 83) and validation (n = 221) cohorts comprising donors (n = 11+26) and patients with chronic pancreatitis (CP, n = 11+20) or tumors: i) benign (serous cystadenoma SCA, n = 13+20), ii) premalignant (intraductal papillary mucinous neoplasms with low/intermediate-grade dysplasia (IPMNdys, n = 8+36) and with high-grade dysplasia/carcinoma in situ (IPMNtis, n = 1+19)), and iii) malignant (IPMNs with an associated invasive carcinoma (IPMNinv, n = 4+14) and ductal adenocarcinomas including anaplastic (n = 4+7), adenosquamous (n = 4+11) and PDAC (n = 27+68)). Pancreatic pCSPG4 expression was evaluated using qRT-PCR (n = 139), western blot analysis and immunohistochemistry. The patients’ characteristics are given in Tables 1 and 2.
Table 1. Characterization of the patients and data summary.
| Test cohort | Validation cohort | |||||||
| Pancreas pCSPG4 [mRNA/10 kCPB] n = 139 | Sera sCSPG4[ng/ml] n = 83 | Sera sCSPG4[ng/ml] n = 221 | ||||||
| Diagnosis | Condition | Age, yearsmedian [range] | No. patients (female/male) mean±SEM median [IQR] | |||||
| Donor | Norm | Test: 44 [16–71]Valid: 43 [19–74] | n = 14 (5/9)12±113 [9]–[16] | n = 116.8±1.74.8 [2.5–11.4] | n = 26 (15/11)7.3±0.66.6 [5.2–8.4] | |||
| CP, chronic pancreatitis | Inflam-mation | Test: 44 [16–76]Valid: 61 [24–78] | n = 15 (3/12)25±326 [14]–[33] | n = 116.8±3.92.8 [1.4–6.1] | n = 20 (7/13)5.0±0.93.7 [2.6–6.5] | |||
| SCA, serous cystadenoma | Benign neoplasm | Test: 61 [38–79]Valid: 60 [44–79] | n = 13 (10/3)159±42113 [37–244] | n = 132.6±0.52.0 [1.9–2.8] | n = 20 (13/7)6.8±1.04.0 [2.3–6.7] | |||
| Intraductal papillary mucinous neoplasms (IPMN) | ||||||||
| IPMNdys, Low- and intermediate-grade dysplasia | Pre-malignant | Test: 60 [41–73]Valid: 65 [42–83] | n = 12 (3/9)20±519 [4]–[29] | n = 84.1±0.94.6 [1.5–5.5] | n = 36 (18/18)4.6±0.34.6 [3.1–5.9] | |||
| IPMNtis, High-grade dysplasia/carcinoma in situ | Pre-malignant | Test: 74Valid: 65 [41–79] | n.d. | n = 1(−/1)1.8– | n = 19 (8/11)3.7±0.52.8 [2.1–4.8] | |||
| IPMNinv, with an associated invasivecarcinoma | Malignant | Test: 66 [46–76]Valid: 64 [46–73] | n = 13 (1/12)29±429 [21]–[39] | n = 41.3±0.31.3 [0.7–1.9] | n = 14 (3/11)7.6±1.27.9 [3.9–9.7] | |||
| Ductal adenocarcinomas (DACs) | ||||||||
| AdSq, adenosquamous carcinoma | Malignant | Test: 65 [51–76]Valid: 69 [41–76] | n = 17 (8/9)39±539 [12]–[49] | n = 46.1±3.13.8 [1.9–12.5] | n = 11 (5/6)3.8±0.53.5 [2.6–5.3] | |||
| Anaplastic, undifferentiatedcarcinoma (G4) | Malignant | Test: 56 [45–74]Valid: 66 [53–75] | n = 10 (4/6)54±2731 [26]–[45] | n = 45.0±2.04.3 [1.7–9.1] | n = 7 (3/4)6.0±1.44.6 [2.7–11.1] | |||
| PDAC (G1–G3), pancreatic ductaladenocarcinoma | Malignant | Test: 66 [38–80]Valid: 65 [39–81] | n = 45 (20/25)36±1116 [8]–[39] | n = 272.6±0.41.5 [1.1–4.1] | n = 68 (30/38)5.2±0.73.5 [1.9–6.3] | |||
Table 2. Circulating sCSPG4 in pancreatic diseases: ROC curve analyses.
| Group | Test cohort | Validation cohort | ||||||||
| n = 83 | median | AUC | 95% CI | P-value | n = 221 | median | AUC | 95% CI | P-value | |
| Donor | 11 | 4.8 | 26 | 6.6 | ||||||
| vs. | ||||||||||
| Chronic pancreatitis | 11 | 2.8 | 0.682 | 0.451–0.912 | 0.149 | 20 | 3.7 | 0.779 | 0.631–0.927 | 0.001 |
| SCA | 13 | 2.0 | 0.811 | 0.629–0.992 | 0.010 | 20 | 4.0 | 0.760 | 0.606–0.913 | 0.003 |
| IPMNdys | 8 | 4.6 | 0.619 | 0.359–0.880 | 0.386 | 36 | 4.6 | 0.787 | 0.677–0.898 | <0.0001 |
| IPMNtis | [1] | – | – | – | – | 19 | 2.8 | 0.883 | 0.766–0.990 | <0.0001 |
| IPMNinv | [4] | – | – | – | – | 14 | 7.9 | 0.515 | 0.299–0.732 | 0.876 |
| IPMNtis+inv | 5 | 1.3 | 0.927 | 0.784–1.070 | 0.008 | 33 | 4.1 | 0.727 | 0.593–0.861 | 0.003 |
| AdSq DAC | 4 | 3.8 | 0.546 | 0.198–0.893 | 0.794 | 11 | 3.5 | 0.862 | 0.735–0.986 | <0.001 |
| Anaplastic DAC (G4) | 4 | 4.3 | 0.636 | 0.304–0.970 | 0.434 | 7 | 4.6 | 0.670 | 0.386–0.955 | 0.172 |
| PDAC (G1–G3) | 27 | 1.5 | 0.784 | 0.623–0.945 | 0.007 | 68 | 3.5 | 0.768 | 0.675–0.860 | <0.0001 |
| - Early PDAC (St. Ia-IIa) | 3 | 0.6 | 0.818 | 0.501–1.136 | 0.102 | 12 | 2.0 | 0.914 | 0.763–1.064 | <0.0001 |
| - Late PDAC (St. IIb-IV) | 24 | 1.5 | 0.783 | 0.612–0.953 | 0.009 | 56 | 3.9 | 0.736 | 0.631–0.841 | <0.0001 |
| all IPMNs | 13 | 1.9 | 0.738 | 0.530–0.946 | 0.049 | 69 | 4.5 | 0.758 | 0.663–0.854 | <0.0001 |
| all DACs1 | 35 | 1.9 | 0.740 | 0.575–0.906 | 0.017 | 86 | 3.6 | 0.772 | 0.688–0.856 | <0.0001 |
| all malignancies2 | 39 | 1.8 | 0.764 | 0.605–0.921 | 0.008 | 119 | 3.6 | 0.764 | 0.683–0.836 | <0.0001 |
| all neoplasms3 | 61 | 2.0 | 0.755 | 0.600–0.911 | 0.007 | 175 | 3.8 | 0.765 | 0.694–0.836 | <0.0001 |
| all pancreatic disorders | 72 | 2.0 | 0.748 | 0.589–0.899 | 0.001 | 195 | 3.8 | 0.767 | 0.698–0.835 | <0.001 |
| Chronic pancreatitis | 11 | 2.8 | 20 | 3.7 | ||||||
| vs. | ||||||||||
| SCA | 13 | 2.0 | 0.584 | 0.334–0.833 | 0.487 | 20 | 4.0 | 0.502 | 0.320–0.685 | 0.978 |
| IPMNdys | 8 | 4.6 | 0.563 | 0.295–0.830 | 0.650 | 36 | 4.6 | 0.549 | 0.379–0.717 | 0.550 |
| IPMNtis | [1] | – | – | – | – | 19 | 2.8 | 0.607 | 0.425–0.789 | 0.255 |
| IPMNinv | [4] | – | – | – | – | 14 | 7.9 | 0.732 | 0.552–0.911 | 0.023 |
| IPMNtis+inv | 5 | 1.3 | 0.773 | 0.541–1.004 | 0.089 | 33 | 4.1 | 0.537 | 0.378–0.695 | 0.653 |
| PDAC | 27 | 1.5 | 0.620 | 0.415–0.823 | 0.253 | 68 | 3.5 | 0.563 | 0.434–0.692 | 0.392 |
| - Early PDAC (St. Ia-IIa) | 3 | 0.6 | 0.712 | 0.326–1.098 | 0.276 | 12 | 2.0 | 0.752 | 0.564–0.939 | 0.019 |
| - Late PDAC (St. IIb-IV) | 24 | 1.5 | 0.619 | 0.407–0.830 | 0.270 | 56 | 3.9 | 0.523 | 0.386–0.659 | 0.764 |
| all IPMNs | 13 | 1.9 | 0,566 | 0.328–0.805 | 0.582 | 69 | 4.5 | 0.543 | 0.397–0.689 | 0.559 |
| all DACs1 | 35 | 1.9 | 0.567 | 0.367–0.765 | 0.511 | 86 | 3.6 | 0.545 | 0.420–0.672 | 0.534 |
| all malignancies2 | 39 | 1.8 | 0.592 | 0.395–0.788 | 0.354 | 119 | 3.6 | 0.522 | 0.399–0.645 | 0.753 |
| all neoplasms3 | 61 | 2.0 | 0.570 | 0.374–0.766 | 0.462 | 175 | 3.8 | 0.505 | 0.380–0.630 | 0.938 |
| IPMNdys | 8 | 4.6 | 36 | 4.6 | ||||||
| vs. | ||||||||||
| IPMNtis+inv | 5 | 1.3 | 0.850 | 0.638–1.062 | 0.041 | 33 | 5.4 | 0.528 | 0.386–0.670 | 0.692 |
| IPMNtis | – | – | – | – | – | 19 | 2.8 | 0.689 | 0.534–0.843 | 0.022 |
| IPMNinv | – | – | – | – | – | 14 | 7.9 | 0.691 | 0.507–0.874 | 0.038 |
| IPMNtis | [1] | 19 | 2.8 | |||||||
| vs. | ||||||||||
| IPMNinv | [4] | – | – | – | – | 14 | 7.9 | 0.791 | 0.631–0.952 | 0.005 |
Grouping of the tumors in: 1 (all DACs): AdSq, Anapl, PDAC; 2 (all malignanices): AdSq, Anapl, PDAC, IPMNinv; 3 (all neoplasms): SCA, IPMNs, DACs.
Cell Cultures, Media, Antibodies
Nine DSMZ-certified pancreatic cancer cell lines (Aspc1, Bxpc3, Capan1, Colo357, HS766T, MiaPaca2, Panc1, SU8686, T3M4) and the cervical carcinoma HeLa cell line (positive control for western blot and ELISA analyses) were cultured in RPMI medium supplemented with 10% fetal bovine serum (FBS; PAA Laboratories GmbH, Pasching, Austria). Primary pancreatic stellate cells (PSC) were obtained through the outgrowth method of Bachem et al. [22], cultured in low glucose DMEM/F12 (1:1) medium supplemented with 20% FBS and propagated for up to 8 passages as previously described [23], [24]. Immortalized human pancreatic ductal epithelial cells (HPDE) were received as a gift [25], and cultured in serum-free keratinocyte medium, supplemented with 5 ng/ml recombinant epidermal growth factor (rEGF) and 50 µg/ml bovine pituitary extract (Life Technologies GmbH, Darmstadt, Germany).
The panel of primary antibodies included the mouse monoclonal anti-CSPG4 antibody raised against melanoma cells (LHM2 clone; MAB2585; R&D Systems/RnD, Minneapolis, MN, USA), the rabbit polyclonal anti-CSPG4 antibody raised against recombinant core protein (H-300; sc-20162; Santa Cruz Biotechnologies/SCBT, Santa Cruz, CA, USA), and the mouse anti-collagen VI (COL6) antibody (64CH11 clone, ab49273; Abcam, Cambridge, UK). The secondary antibodies and isotype controls used for immunoblotting, immunohistochemistry, immunofluorescence, and FACS analyses are indicated in the respective sections.
siRNA Transfection and Functional Studies
To confirm the specificity of the anti-CSPG4 antibodies and to evaluate the functional relevance of pCSPG4 in pancreatic cancer, we used siRNA-based knock-downs. Cells were grown up to 50–70% confluence and transfected using the HiPerFect transfection reagent (Qiagen GmbH, Germany) at 10 nM with duplex oligonucleotides: siRNA set 1 (sense: CGG UGA GGG AUG UAA AUG Att; antisense: UCA UUU ACA UCC CUC ACC Gtg), siRNA set 2 (sense: AAA UCU CCG UGG ACC AGU Att; antisense: UAC UGG UCC ACG GAG AUU Ucc) (Ambion Applied Biosystems, USA), or control siRNA set (AAT TCT CCG AAC GTG TCA CGT) (Qiagen, Hilden, Germany) for 48 hours.
To evaluate the effect of CSPG4 gene silencing on cell functions, proliferation, migration, and invasiveness, the cells were treated with control or CSPG4-specific siRNA and analyzed through MTT-based growth assay, scratch test, and Matrigel-based invasion assay, using the standard techniques reported elsewhere [23], [24].
Induction of Hypoxia
Pancreatic cell lines were grown up to 70% confluence, transferred to the modular incubator chamber (Billups-Rothenberg Inc., Del Mar, CA, USA), flashed for 30 min with the hypoxic gas mixture (0.74% O2 and 9.9% CO2 in N2), and incubated in the closed unit for 3 h or 48 h at 37°C. The same procedure was performed without exposure to hypoxic gas to obtain a normoxic control.
FACS Analysis
Pancreatic cell lines were suspended in FACS Buffer (2% FBS in PBS), blocked with FcR Blocking Reagent (Miltenyi Biotech GmbH, Begrisch Gladbach, Germany), and incubated for 20 min with mouse anti-CSPG4 antibody (LHM2 clone; RnD) at room temperature, or IgG1 isotype control (SCBT). We used directly labeled phycoerythrin (PE)-conjugate or unlabeled antibody with subsequently added anti-mouse AlexaFlour488-conjugate. Measurements of expression under normoxic and hypoxic conditions were performed using the FACScan and LSR flow cytometers (Becton Dickinson & Co, Franklin Lakes, NJ, USA).
Enzyme-Linked Immunosorbent Assay (ELISA)
Determination of the total CSPG4 protein in biofluids (sera and media conditioned by cell lines) was performed using commercial quantitative sandwich enzyme immunoassays (ELISA kits). The test cohort was evaluated in 2011–2012 using an ELISA kit manufactured by USCN Life Science Inc. (Wuhan, P. R. China; Human Melanoma Associated Chondroitin Sulfate Proteoglycan/MCSP assay; Cat.# E91134Hu). This company became part of the Cloud-Clone Corp. (Houston, TX, USA) and continued to sell the kit under Cat.# SEB134Hu. It should be noted that Cloud-Corp revised the instruction manual for this kit as of August 12, 2013, relabeling the vial with the standard stock as 100 ng/ml, rather than the 10 ng/ml previously given, and introducing a 1:10 dilution step to obtain 10 ng/ml to be used for consecutive serial 1:2 dilutions. We purchased this kit for independent validation studies in October–December 2013 and first performed control retesting of randomly chosen ‘old’ samples. This procedure delivered log-higher sCSPG4 values, i.e., 5 ng/ml instead of 0.5 ng/ml. Therefore, for presentation, we re-scaled all original measurements of the test cohort by a factor of 10.
As a control, we i) confirmed sCSPG4-identity of the standard by performing western blot analysis with H-300/SCBT antibody (not shown), ii) proved the titratability of the sera samples (not shown), and iii) validated the findings with a different ELISA kit (results in the main text). For the latter, we randomly choose samples to represent each tested group and measured sCSPG4 in 64 sera with a commercial kit manufactured by CUSABIO (CSB-EL006076HU; Wuhan, PRC). The USCN/Cloud-Clone kit employed a recombinant N-terminal fragment of a protein (Leu16-Ser350) as an ELISA standard and immunogen to produce mouse monoclonal (capture) and goat polyclonal (detection) antibodies; CUSABIO - a full-length protein expressed in eukaryotic cells. To enable colorimetric reaction, both USCN/Cloud-Clone and CUSABIO kits used avidin-labeled detection antibodies, streptavidin-HRPO conjugate, and TMB substrate. HeLa-supernatant served as positive control for both ELISAs and delivered values of 11.0 ng/ml and 6.0 pg/ml for Cloud-Clone (standard range: 0–10 ng/ml) and CUSABIO (standard range: 0–1600 pg/ml) immunoassays, respectively. Thus, both ELISAs detected a native human sCSPG4 ectodomain, albeit most probably its different epitopes.
Expression Profiling of Tissues
Sentrix Human-6v3 Whole Genome Expression BeadChips (Sentrix Human WG-6; Illumina) were used to identify genes expressed in pancreatic cancer tissue. To synthesize first and second strand cDNA and amplify biotinylated cRNA from the total RNA we used an Illumina TotalPrep RNA Amplification Kit. Hybridization to the BeadChip was performed without modification according to the manufacturer’s instructions. A maximum of 10 µl cRNA was mixed with a 20 µL GEX-HYB hybridization solution. The preheated 30 µl assay sample was dispensed onto the large sample port of each array and incubated for 18 hours at 58°C. Following hybridization, the samples were washed according to the protocol and scanned with a Bead Array Reader (Illumina, San Diego, CA). Raw data were exported from the Bead studio software to R. The data were quantile normalized and log2 transformed. The data have been uploaded to ArrayExpress under accession number E-MTAB-1791.
Real-time Quantitative Polymerase Chain Reaction
All reagents and equipment for mRNA/cDNA preparation were supplied by Roche Applied Science (RAS, Mannheim, Germany) [23], [24], [26]. mRNA was prepared by automated isolation using a MagNA Pure LC instrument and isolation kit. cDNA was prepared using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) according to the manufacturer’s instructions. Real-time PCR was performed with the LightCycler and CSPG4 kit supplied by Search-LC (Heidelberg, Germany). The number of CSPG4 transcripts in the samples was normalized to the expression of the housekeeping gene, cyclophilin B. The final data are presented as the number of transcripts per 10,000 CPB copies (10 kCPB).
Western Blot Analysis (Immunoblotting)
Cells and tissues were lysed in RIPA-Buffer supplemented with proteinase inhibitors (complete mini, RAS) [23]. 30 µg-protein samples were separated using the NuPAGE Gel system (4-12% Tris-Glycine gel; Invitrogen, Karlsruhe, Germany) and transferred to a nitrocellulose membrane (BioRad, Munich, Germany). Blots were blocked with 5% milk powder in TBS, incubated with mouse LHM2 or rabbit H-300 anti-CSPG4 antibodies overnight at 4°C, followed by the peroxidise-conjugated secondary anti-mouse or anti-rabbit antibodies for 1 hour, and exposed to a chemiluminescence detection kit (all reagents from ECL Amersham; GE Healthcare Europe GmbH, Freiburg, Germany). For equal loading control analysis, the membrane was exposed to stripping buffer (Thermo Scientific, Rockford, IL, USA) for 30 min at 37°C, and then incubated with an antibody against GAPDH (36 kDa) or beta-actin (42 kDa) (SCBT). Commercial SK-MEL-28 melanoma cell lysate (SCBT) and freshly prepared HeLa cells lysate were used as positive controls for CSPG4 detection.
Immunohistochemistry
Three micrometer-thick paraffin-embedded tissue sections were immunostained using a peroxidase-labeled polymer and Envision detection system (Dako GmbH, Hamburg, Germany) to visualize the binding of primary antibodies [3], [23]. Briefly, antigens were retrieved by boiling the tissue sections in 10 mM citrate buffer for 10 min in a microwave oven, then the endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 10 min, and the nonspecific binding was blocked with Power Block (BioGenex Laboratories, Fremont, CA, USA). Sections were incubated with mouse LHM2 (RnD) or rabbit H-300 (SCBT) anti-CSPG4 antibodies at 4°C overnight. Normal mouse IgG1s or rabbit IgGs (Dako) were used as negative controls. The HRP-labeled anti-mouse or anti-rabbit polymers (Dako) were applied at room temperature for 45 min. The reaction product was visualized using a DAB chromogen/H2O2 substrate mixture, resulting in brown staining, and sections were counterstained with Mayer’s haematoxylin. Slides were analyzed using an Axioplan-2 imaging microscope (Carl Zeiss Microscopy GmbH, Jena, Germany).
Immunofluorescence and Confocal Laser Microscopy
Paraformaldehyde-fixed tissues and cell lines additionally treated with 0.3% Triton X-100 for 5 min were incubated with blocking solution (2% FBS, 2% bovine serum albumin, 0.2% gelatine in PBS), and exposed to mouse LHM2 anti-CSPG4 antibody (single staining) or to a combination of the rabbit anti-CSPG4 (H-300) and mouse anti-COL6 (64CH11) antibodies (double staining). After overnight incubation at 4°C, the secondary antibodies were applied at room temperature for 1 h: anti-mouse AlexaFluor488 (green)-conjugated IgG (single staining) or anti-rabbit Cy2 (green)-conjugate and anti-mouse Cy3 (red)-conjugate (double staining). Normal mouse IgG1 and IgG2a or rabbit IgGs were used as negative controls. The nuclei were visualized through DAPI. Slides were analyzed using an Axioplan-2 imaging microscope. Double stained slides were also viewed with a confocal laser scanning microscope (TCS-SP, Leica Microsystems GmbH, Wetzlar, Germany; courtesy of Dr. N. Brady; Bioquant/DKFZ).
Statistical Analysis
Statistical analyses and graphical data presentation were performed using GraphPad Prism 5 (Graph Pad Software, La Jolla, CA). The quantitative variables are presented as dots showing raw values and bars showing the median and interquartile range (IQR). The significance of differences between groups was assessed using non-parametric methods (Mann-Whitney test for two groups and Kruskal-Wallis with Dunn’s test for multiple groups). CSPG4-underexpression was determined as a negative value obtained upon log2-transformation of the individual measurements first normalized to the total median level. The respective median levels are depicted as green lines in Figs. 1A, 1D and 2A, and show 2.3 ng/ml for the sCSPG4 test cohort (n = 83), 4.2 ng/ml for the sCSPG4 validation cohort (n = 221), and 24 copies/10 kCPB for the pCSPG4 mRNA cohort (n = 139). The discriminatory potential of the serum sCSPG4 was determined by means of receiver operating characteristic (ROC) curves. The regression analyses were performed using the IBM SPSS19 statistics software (IBM Co, Armonk, NY, USA). The overall survival rate from the date of initial surgery was estimated using the Kaplan–Meier method and differences between survival curves were analyzed using the log-rank test. Two-sided P values were computed and a difference was considered statistically significant at p≤0.05 (*), p<0.01(**) or p<0.001(***).
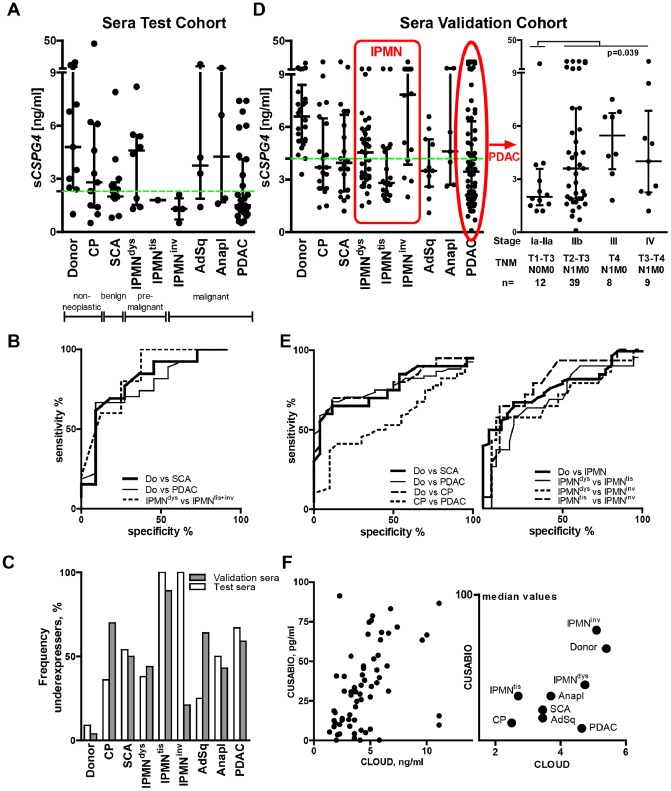
Figure 1. Reduction of serum CSPG4 (sCSPG4) in pancreatic diseases and its selective preservation following the ‘drop and restoration’ pattern in advanced IPMN and PDAC.
(A) Systemic levels of sCSPG4 were determined by ELISA in sera (USCN/Cloud-Clone Corp. kit; n = 83) of healthy volunteers and patients with chronic pancreatitis (CP), serous cystadenoma (SCA), premalignant (dysplastic IPMNdys and IPMNtis), and malignant (IPMN with an associated invasive carcinoma, IPMNinv) forms of intraductal papillary-mucinous neoplasm, as well as in ductal (PDAC), adenosquamous (AdSq) and anaplastic (Anapl) carcinomas. The data are summarized to show individual values, median level, and interquartile range (IQR). (B) The ROC analyses of the ELISA data showed the discriminatory power of sCSPG4 levels for patients with different pancreatic diseases. The dotted green line represents the median level of the particular cohort used to estimate the frequency of the sCSPG4 underexpressers in the panel (C), as detailed in Materials and Methods. (D–E) Validation of ELISA findings in an independent cohort (n = 221). Red-bordered areas indicate subgroups depicting IPMN and PDAC progression. The patients’ characteristics and results of statistical analyses are presented in the main text and Tables 1 and 2. (F) Validation of ELISA findings with a different ELISA kit (CUSABIO; n = 64).
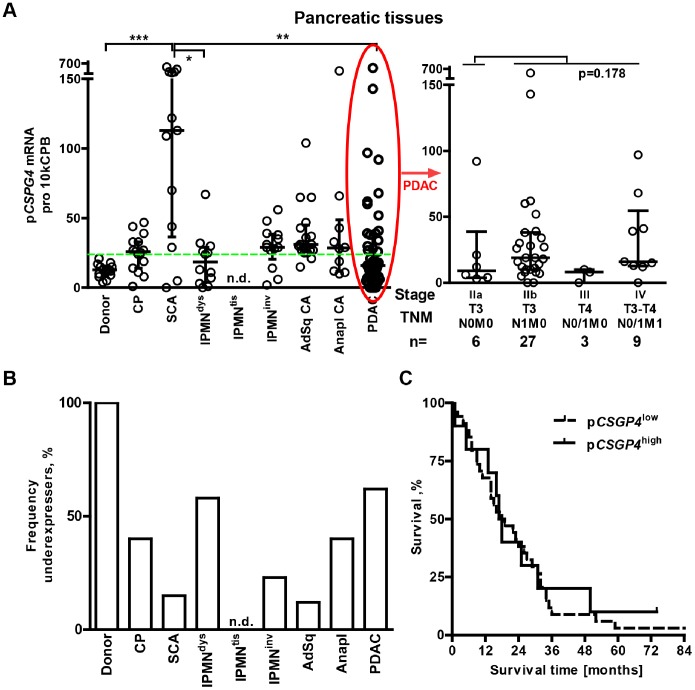
Figure 2. Maintenance of pCSPG4 mRNA expression in pancreatic tissues.
(A) The pCSPG4 copy number was measured using qRT-PCR in pancreatic tissues (n = 139) and normalized to that of the housekeeping gene cyclophilin B (CPB). It is presented as the amount of transcripts per 10 kCPB copies. An elevated pCSPG4 mRNA level was observed in all groups compared to donors (p-values <0.01; Mann-Whitney tests), except IPMNdys (p = 0.47) and PDAC (n = 0.20). Kruskal-Wallis and Dunn’s test analysis, comparing multiple groups within the entire cohort, established an overexpression of pCSPG4 in serous cystadenoma, SCA (*p<0.05, **p<0.01 and ***p<0.001 significance level). The dotted green line represents the median level used to estimate the frequency of the underexpressers in the panel (B). (C) Kaplan-Meier survival curves for pCSPG4 low and pCSPG4 high (upper quartile; over 38 copies/10 kCPB) groups.
Results
Preferential Reduction of Serum CSPG4 (sCSPG4) in Pancreatic Diseases
We suggested that extracellular shedding of CSPG4 might normally occur in human tissues and lead to the release of the soluble ectodomain (sCSPG4) into circulation. Thus, propagation of CSPG4-expressing pericytes and tumor cells might suffice to raise systemic levels of sCSPG4 in patients with malignancies. Using an ELISA kit (USCN/Cloud-Clone Corp.), we tested sera of healthy volunteers (n = 11) and patients with the four most common pancreatic diseases (n = 72): chronic pancreatitis (CP), cystadenoma (SCA), intraductal papillary mucinous neoplasms (IPMNdys, IPMNtis, and IPMNinv with low/intermediate-, high-grade dysplasia, and with invasive carcinomas), and ductal malignancies (PDAC, adenosquamous and anaplastic) (Table 1). This first-ever evaluation of circulating sCSPG4 established that the normal median concentration was 4.8 ng/ml, and the mean 6.8 ng/ml. Unexpectedly, sCSPG4 levels were lower in sera of patients with pancreatic disorders (Fig. 1A; p = 0.01). The drop in sCSPG4 had already been seen in the inflammatory CP group (n = 11), but that altered level did not show a statistically significant difference from either normal (p = 0.158) or neoplastic ones (p-values ranging from 0.100 to 0.690). sCSPG4 reduction reached significance in the neoplastic group (n = 61, p = 0.008), mostly due to a remarkable decline in patients with benign SCA (n = 13, p = 0.011), IPMNtis+inv carcinomas (n = 5, p = 0.009), and PDAC (n = 27, n = 0.007). The sCSPG4 differences observed discriminated these patients from donors–but not CP patients–by means of ROC curve analysis (Fig. 1B; Table 2, test cohort). Notably, patients’ sCSPG4 values were not ubiquitously down-regulated; some patients appeared to maintain normal sCSPG4 levels while an increased number presented extremely low values, raising the frequency of underexpressers (Fig. 1C).
Remarkably, IPMN carcinomas (IPMNtis+inv; one case of carcinoma in situ/Tis and four cases of invasive tumors) were associated with the strongest drop in sCSPG4 (p = 0.009), whereas premalignant IPMNdys cases showed the best level of sCSPG4 retention (n = 8, p = 0.471). The sCSPG4-distinction among IPMNs (IPMNdys vs. IPMNtis+inv, p = 0.047) appeared to suffice for ROC discrimination of these entities (Fig. 1B; Table 2, test cohort). This indication, however, should be viewed with caution due to the inability of such a small cohort to represent heterogeneity of IPMNs [27]–[30].
Among ductal malignancies (n = 35, p = 0.018), the sCSPG4 drop was more prominent in patients with differentiated adenocarcinoma (PDAC G1-G3: n = 27, n = 0.007) than in less differentiated anaplastic (G4: n = 4, p = 0.473) and adenosquamous (n = 4, p = 0.845) variants. Nevertheless, the degree of tumor cell differentiation did not determine sCSPG4 variance (p = 0.125 and p = 0.148 compared to PDAC, respectively; p = 0.40 for PDAC G2 vs. G3). In contrast, sCSPG4 in sera of patients with metastatic PDAC (UICC Stage IV: n = 5 N1M1, p = 0.610) was significantly higher than in those with non-metastatic PDAC (UICC Stages I–IIb: n = 22, with 3 N0M0 and 19 N1M0, p = 0.003), without exceeding normal amounts. sCSPG4 level in PDAC, however, did not have prognostic relevance (survival time analyses using Cox regression [p = 0.509] and the Kaplan-Meier method comparing sCSPG4 low and sCSPG4 high>Q3 groups with median survival times of 20 and 22 months, log-rank test p = 0.450).
Thus, the test cohort data supported our hypothesis that sCSPG4 would be found in circulation. It also indicated that the development of pancreatic diseases may reduce systemic sCSPG4 levels. Seemingly normal levels in certain subgroups of the patients could be attributable either to patient-specific preservation or to cell/stage-specific restoration. The latter variant means that in pancreatic disease, sCSPG4 changes might generally follow a ‘drop and selective restoration’ pattern.
Independent Validation of the ‘Drop and Restoration’ Pattern
Our test cohort represented a selection of 72 patients affected by the four most common pancreatic diseases. As a small sample size can lead to overstatements, we analyzed sCSPG4 in an independent cohort (n = 221) consisting of 26 donors and 195 patients (Figs. 1D–F, Tables 1 and 2, validation cohort). The normal median level was 6.6 ng/ml and the mean 7.3 ng/ml (no significant difference from the test cohort donors according to the Mann-Whitney test, p = 0.240). Independent validation established the reduction of circulating sCSPG4 as a statistically significant feature of all pancreatic diseases, including CP (3.7 ng/ml, n = 26, p<0.001).
We took particular care to prove the ‘drop and restoration’ assumption by increasing the number of patients representing different stages of PDAC and IPMN progression. Raising the IPMN cases from 13 (8dys, 1tis, and 4inv) in the test cohort to 69 (36dys, 19tis, and 14inv) in the validation cohort substantiated the sCSPG4-distinctivity of IPMNs and highlighted the heterogeneity of IPMNtis+inv carcinomas. Taken as separate entities, non-invasive IPMNtis showed the lowest observed level of sCSPG4 (2.8 ng/ml), whereas invasive IPMNinv displayed the highest (7.9 ng/ml). Although the validation study relocated non-invasive IPMNdys with low−/intermediate-grade dysplasia from the ‘preserved‘ to the ‘reduced’ position (4.6 ng/ml, n = 36, p = 0.0001), the level was still higher than in IPMNtis with high-grade dysplasia (p = 0.023), but lower than in IPMNinv with associated invasive carcinoma (p = 0.039). These differences confirmed the ROC curve-based discrimination of IPMN entities from the donors as well as from each other (Fig. 1E and Table 2, validation cohort). However, the previously observed distinction between IPMNdys (high) and IPMNtis+inv (low) should preferentially be attributed to the non-invasive IPMNtis group. Therefore, the discrepancy between the IPMNinv profile in the test and validation cohorts (Figs. 1A, C and D) demonstrated most strongly how sample size might impact the conclusion, particularly when studying a disease the morphological classification of which alone does not fully encompass its complexity or heterogeneity, and possibly distinct carcinogenic pathways [27]–[31]. Nevertheless, taken as an adenoma–carcinoma sequence, IPMN’s dys→tis→inv progression appeared to deliver a ‘drop and restoration’ sCSPG4 pattern, with progressive reduction in the circulation of sCSPG4 in premalignant stages and compensatory gain in the advanced stage.
In PDAC (n = 68), the reduced sCSPG4 levels were not distinguishable from those in CP (p = 0.394). However, the PDAC subgroup with early disease (lymph node and/or metastasis-free) showed the lowest median sCSPG4 value (2 ng/ml; IUCC stages Ia-IIa: T1-3N0M0, n = 12; Fig. 1D, right panel). The value for this subgroup was significantly lower than that for the CP group (n = 0.019), and also the late/advanced PDAC group (3.9 ng/ml; stages IIb–IV: T2-4N1M0-1, n = 56, p = 0.0386). Here, the impact of the nodal but not metastatic status reached statistical significance (for 59 M0 vs. 9 M1 patients, p = 0.520). As in the test cohort, the degree of tumor cell differentiation did not determine sCSPG4 variance (PDAC G2 vs. G3, p = 0.23; PDAC, anaplastic and adenosquamous variants, p = 0.372).
Overall, the validation study reinforced the positioning of any pancreatic disease as an sCSPG4-reducing factor, with certain (aggressive/advanced) malignancies possessing sCSPG4-restoring features.
Validation of the Findings with a Different ELISA Kit
The observed reduction in the sCSPG4 level could be absolute or relative, i.e., reflecting either the disappearance of proteoglycans from circulation entirely or of recognizable epitopes from the still circulating sCSPG4. To address this issue, we re-measured the validation sera with a different ELISA kit. Instead of the N-terminal (Leu16-Ser350) fragment used by USCN/Cloud-Clone Corp., this CUSABIO-manufactured kit employed full-length immunogen to generate monoclonal capture and polyclonal detection antibodies. In the control experiment, both ELISAs measured sCSPG4 shed in the supernatants by HeLa cells, albeit at different levels: 11 ng/ml by Cloud-Clone and 6 pg/ml by CUSABIO. The sera data also showed ng and pg levels of measurements, with significant correlation of individual sCSPG4 values in 64 randomly chosen samples (Fig. 1F, left panel; Spearman Rho = 0.52; p<0.0001). Moreover, the plotting of median values confirmed the sCSPG4-reducing character of pancreatic diseases (Fig. 1F, right panel). These data suggest that the disappearance of sCSPG4 from circulation is more likely to occur than a loss of specific epitopes.
Such a drop in circulating sCSPG4 could be the result of excess degradation or reduced production, occurring more frequently in pancreatic patients than in healthy individuals. A few samples showing discrepant CUSABIOhigh/Cloudlow or CUSABIOlow/Cloudhigh patterns contradicted the common degradation of circulating sCSPG4 in pancreatic diseases. Furthermore, in the case of proteolysis, at least one of the cleaved sCSPG4-fragments should be recognized by an antibody at the non-cleaved-equivalent level (see Discussion). Alternatively, the reduction of systemic sCSPG4 could reflect diminished production and/or release in different organs, including a disorganized pancreas.
Differential Expression of pCSPG4 in Human Pancreatic Tissues (pCSPG4)
To clarify whether observed drops in circulating sCSPG4 levels were due to reduced intrapancreatic production (pCSPG4), we evaluated pCSPG4 mRNA expression in normal (organ donor), inflammatory (chronic pancreatitis), and neoplastic pancreata using qRT-PCR (Table 1 and Fig. 2). Surprisingly, the reduction in serum protein levels was contrasted by maintained or elevated pancreatic levels (Fig. 2A). Median pCSPG4 mRNA values were higher in all patient groups compared to donors, except IPMNdys (n = 12, p = 0.47) and PDAC (n = 45, p = 0.20). The frequency of underexpressers was significantly reduced (Fig. 2B). pCSPG4 mRNA was overexpressed in 5–25% of malignant specimens and was exceptionally high in almost all benign serous cystadenoma (SCA) biopsies. SCA was the only case of a significant 10-fold increase compared to the donor and CP groups, as well as 2- to 5-fold increase compared to other pancreatic neoplasms (p<0.001); it also had the highest pCSPG4 high/sCSPG4 low discordance. pCSPG4 mRNA expression in PDAC lesions did not correlate with any of the clinico-pathological parameters, such as age, sex, tumor grading staging, or survival (not shown). In the most frequently diagnosed PDAC subgroup (n = 44, as one patient was lost for follow-up), the Kaplan-Meier analysis showed similar overall survival rates of patients with low and high (upper quartile; >38 copies/10 kCPB) pCSPG4 expression (Fig. 2C; 18.5 vs. 17.5 months; p = 0.738). Thus, the pCSPG4 pattern differed among pancreatic diseases, and showed diagnostic but not prognostic relevance.
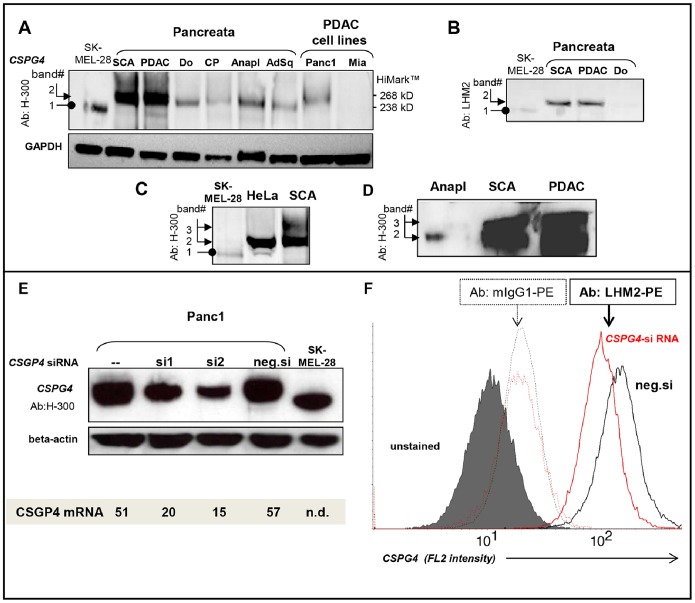
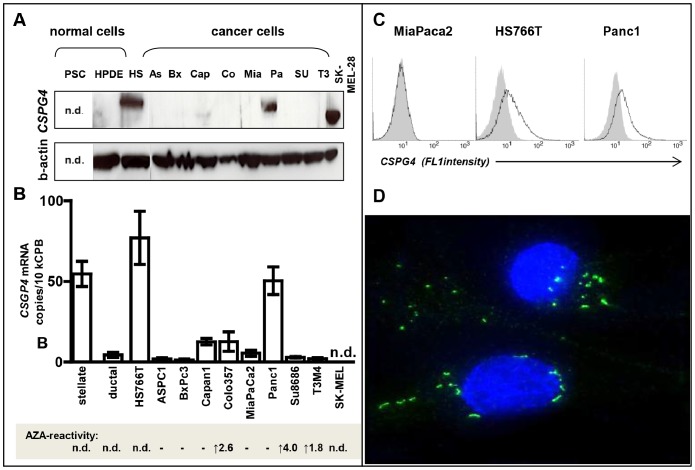
Although pCSPG4 mRNA expression was not reduced, it did not exclude the possibility of decreased protein expression. Both core protein-specific polyclonal rabbit H-300 antibodies (immunogen: E.coli-produced recombinant protein; Fig. 3A) and the surface epitope-specific monoclonal mouse LHM2 antibodies (immunogen: melanoma cells A375P and SK23; Fig. 3B) recognized pCSPG4 protein in pancreatic tissues upon western blot analysis. The molecular weight of normal pancreatic pCSPG4 protein (band-2, ca. 275 kDa) was higher than that of the melanoma antigen used as a positive control (band-1, ca.250 kDa; lysate of SK-MEL-28 cells). Importantly, inflammatory and neoplastic pancreata retained normal band-2, which was also a major product in ELISA-positive HeLa cells (Fig. 3C). The difference from melanoma was similar to that recently described in human fetal brain and glioblastoma specimens [19]. In the PDAC and SCA samples, this band-2 isoform was overexpressed and frequently accompanied by an additional higher-sized product (band-3, ≥300 kDa, Fig. 3D). Western blot and FACS analyses of siRNA-based CSPG4 knockdowns in the Panc1 cell line confirmed the CSPG4 nature of the pancreatic isoform and further validated the pCSPG4-specificity of H-300 and LHM2 antibodies (Figs. 3E–F).
Figure 3. pCSPG4 protein expression in pancreatic tissues.
Immunoblotting confirmed the specificity of polyclonal H-300 (A) and monoclonal LHM2 antibodies (B), generated against recombinant protein and melanoma-derived antigens, respectively. The images demonstrate differences in size between pancreatic (band-2) and melanoma (SK-MEL-28, band-1) antigens, but not cervical carcinoma (HeLa) antigens (C), and reveal the frequent existence of oversized isoforms in SCA and PDAC (band 3, C–D). GAPDH was used as the loading control. HiMark (Invitrogen) was used as the molecular weight marker. (E) Western blot and (F) FACS analyses of CSPG4 siRNA-transfected Panc1 cells further confirmed the specificity of the used antibodies and established the knockdown efficacy of the two siRNA sets (si1 and si2) at approximately 75% after 48 h post transfection, compared to the negative control siRNA (neg.si). QRT-PCR confirmed the efficacy of the knockdowns at the mRNA level.
Localization of CSPG4 in Pancreatic Tissues
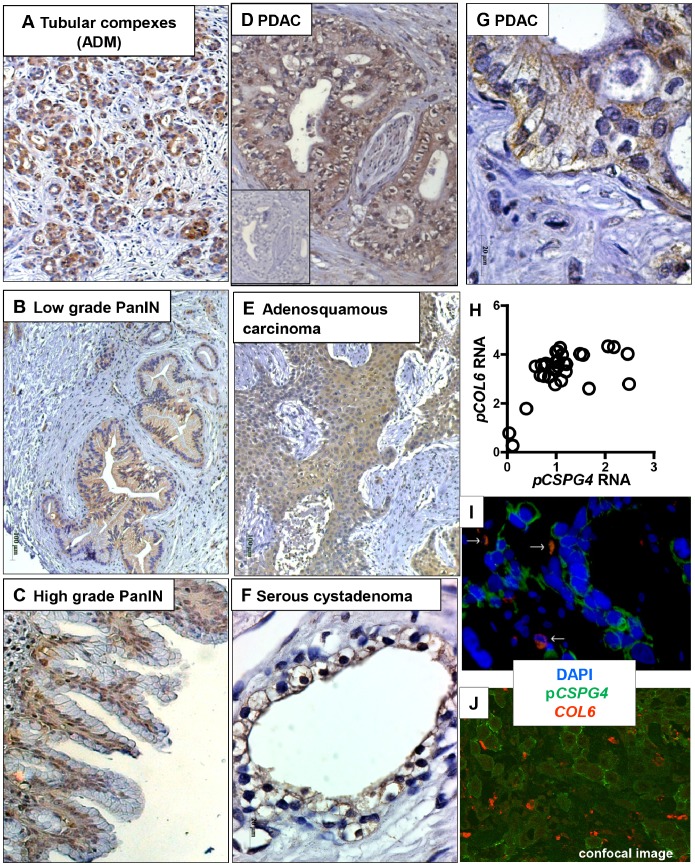
Despite pancreatic disorganization, RNA and the protein levels of pCSPG4 were not reduced in diseased organs. To elucidate the reason for maintained/elevated pCSPG4 expression, we visualized the localization of CSPG4 in pancreatic tissues. The immunohistochemical evaluation of the various specimens revealed that the cells scattered in islets, stroma and the walls of normal ducts or vessels were the common non-malignant source of CSPG4 immunopositivity in normal, inflammatory and cancerous pancreata. A proportion of the cells showed co-immunopositivity for chromogranin A (neuroendocrine origin [10]), as did others for desmin, vimentin and PDGF receptors (fibroblast-like cells of pericyte origin; not shown). Whereas normal pancreatic ducts lacked CSPG4, a strong signal was detected in the tubular complexes emerging among degenerated acini in the paratumoral areas affected by reactive reorganization (Fig. 4A). The same reactive pattern was also found in CP tissues (not shown). Also, premalignant PDAC precursors (PanINs) showed CSPG4 positivity, from weak/diffuse in low-grade PanINs to strong/basal in higher-grade lesions (Fig. 4B–C).
Figure 4. Localization of CSPG4 in pancreatic tissues.
Staining of tissues was performed using mouse LHM2 anti-CSPG4 antibody, followed by HRP-conjugated anti-mouse polymer and visualization with the Dako Envision system (immunohistochemistry, A–G), or using rabbit anti-CSPG4 antibody (H-300) and mouse anti-COL6 antibody followed by anti-rabbit-Cy2 and anti-mouse-Cy3 conjugates (immunofluorescence, I–J). The antigen-specific antibodies were replaced with isotype IgGs for negative control; the representative image is given as inset in (D). Shown (100x–200x) are tubular complexes/acinar-to-ductal metaplasia ADM (A), pre-malignant PanIN lesions of low (B) and high grade (C) in paratumoral areas of PDAC biopsies; perineural invasion of PDAC tumor cells (D); squamous compartment in adenosquamous carcinoma (E); and high-resolution images (630x) of an epithelium lining of cysts in serous cystadenoma, SCA (F) and tumor cells in PDAC lesion (G). (H) Co-expression of CSPG4 and COL6 RNA in pancreatic tissues according to microarray-based measurements. (I) Double immunofluorescent staining showed rare co-localization of pGSPG4 (green) and COL6 (red; white arrows) in PDAC lesions, and prevalence of COL6-free surfaces. The images were routinely recorded using Axiovision Software installed on a Carl Zeiss microscope, and (J) confirmed by confocal laser scanning microscopy (TCS-SP, Leica Microsystems, courtesy of Dr. N. Brady, Bioquant, Heidelberg University/DKFZ, Germany).
The malignant lesions lacked islets (and therefore the corresponding CSPG4 immunoreactivity), but showed irregular focal staining of cancerous ducts in PDAC, strongest in the areas with perineural invasion (Fig. 4D). Diffuse immunopositivity was also observed in squamous elements of adenosquamous carcinoma (Fig. 4E), and in anaplastic carcinomas and invasive IPMN lesions, but not dysplastic IPMN (not shown). Benign SCA showed uniform, prominent accumulation of the CSPG4 in the epithelial lining of the cysts (Fig. 4F).
The staining of epithelium in benign (pCSPG4 high/sCSPG4 low) SCA was exclusively membranous (Fig. 4F); the majority of malignant cells showed diffuse cytoplasmic and/or membranous patterns (Fig. 4G). Type VI collagen (COL6) is not only a major interacting partner but also a powerful blocker of CSPG4 shedding [20]. The microarray-based profiling (Fig. 4H) confirmed the expression of COL6 in malignant pancreatic lesions [32]. Notwithstanding RNA co-expression, double immunofluorescence und confocal microscopy revealed only occasional co-localization of COL6 and pCSPG4, and the prevalence of COL6-free CSPG4-positive interfaces (Fig. 4I, white arrows; 4J, confocal microscopy). sCSPG4 trapping in the extracellular matrix [19] was not observed.
Based on these data, we concluded that the variability of cellular sources allows the pancreas to maintain or elevate pCSPG4 synthesis; certain types of expanding cells, however, may overexpress CSPG4 without shedding it.
Expression and Lack of CSPG4 Release in Pancreatic Cancer Cells
Among pancreatic cancer cell lines, only 3/9 cultures (Panc1, HS766T and, to a much lesser extent, Capan1) showed an accumulation of pCSPG4 protein (Fig. 5A), the molecular weight of which, ca. 275 kDa, was higher than in melanoma (band-1) but similar to that in normal and cancerous pancreata, and in the HeLa cell line (band-2, Figs. 3A and 3C). CSPG4 was not detectable in normal adult ductal cells (HPDE). This pattern was mirrored exactly by the mRNA profile (Fig. 5B), with high constitutive mRNA expression in Panc1 and HS766T comparable to the mRNA level observed in pancreatic stellate cells (PSC) used as a positive control of presumably pericytic origin. In contrast, normal HPDE cells and seven other cancer cell lines showed barely detectable mRNA copies of CSPG4 (a range of 2–7 copies/104 copies of CPB). The CSPG4 silencing was only partially alleviated by DNA methylation inhibitor 5-Aza-2′-Deoxycytidine (Aza); the expression was induced in three out of eight cell lines (Colo357, SU8686, T3M4; average increase of 2.7 times; Fig. 5B, lower panel), thus indicating that hypermethylation is only partially responsible for the low level of CSPG4 transcription in pancreatic tumor cells.
Figure 5. CSPG4 expression in pancreatic tumor cells.
(A) Immunoblotting with H-300 antibody detected pCSPG4 protein in three of nine pancreatic cancer cell lines; shown in relation to melanoma SK-MEL-28 and a normal immortalized ductal epithelial cell line, HPDE. (B) QRT-PCR analysis of pancreatic cancer cell lines, normal epithelial HPDE cells and primary myofibroblasts (pancreatic stellate cell, PSC). (C) Whereas none of the ELISA kits was capable of detecting sCSPG4 in supernatants (not shown), FACS analyses with LHM2 and anti-mouse Cy2-conjugate (bold lines) revealed surficial exposure of CSPG4 by pCSPG4-postive Panc1 and HS766T but not pCSPG4-negative MiaPaca2. (D) The focal accumulation of CSPG4-forming dots and short fibrils was visualized by means of immunofluorescence in Panc1 cells. The specificity of staining was confirmed using mouse isotype IgG1 control (not shown).
FACS analyses confirmed surficial exposure of CSPG4 in Panc1 and Hs766T cells; immunofluorescent staining revealed preferentially fibrilar patterns of CSPG4 distribution (Fig. 5C–D). The secreted/shed form of CSPG4 was undetectable in the supernatants of different pancreatic cancer cell lines, or in highly expressing Panc1 cells (limit of detection 0.15 ng/ml; data not shown) – in contrast to the strong accumulation of sCSPG4 in the supernatants of HeLa cells showing Panc1-like band-2 in the pCSPG4 western blot analysis (Fig. 3C). Apparently, these cells lacked the autocrine shedding mechanism operating in HeLa. Thus, pancreatic cancer cells rarely overexpressed CSPG4 and tended to retain it on the surface without shedding. Therefore, levels of ectodomain in circulation may not necessarily reflect levels of proteoglycan in tissues, and elevation of pCSPG4 will not enforce a rise in sCSPG4.
Functional Relevance of pCSPG4 Expression in Pancreatic Cancer Cells and its Association with Chronic Hypoxia
The lack of prognostic associations, exclusive overexpression of pCSPG4 in benign neoplasm mainly avoiding malignant transformation (SCA), and relatively rare expression of pCSPG4 in carcinoma cell lines argued against the attribution of CSPG4 pro-malignant activity. Thus, the artificial down-regulation of CSPG4 in pCSPG4-expressing cancer cells should not impede their malignant behavior. To prove this, we reduced intrinsic CSPG4 levels by means of two different siRNA sets in Panc1 and MiaPaca2 cells and compared the effects to those seen in control-siRNA treated cultures. Upon knockdown, MiaPaca2 cells showed complete removal of the few existing RNA copies (although both were undetectable by western blot analysis), and Panc1 showed a 70%-ige reduction in the high CSPG4 levels at the RNA and protein levels, total and on the surface (Fig. 3E–F). Proliferation, motility and invasion were neither reduced nor promoted (data not shown), questioning any role of CSPG4 in pancreatic cancer other than as a biomarker.
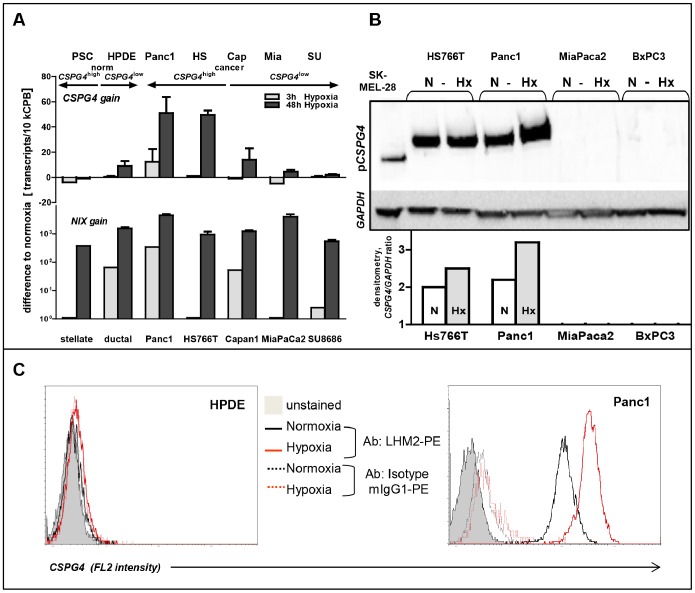
In addition to benignancy, the most striking feature of SCA is its association with disorders caused by gene mutations that affect hypoxic signaling pathways, such as pVHL in von-Hippel-Lindau and LKB1 in Peutz-Jeghers syndromes [33]–[37]. What is particularly frequent is an association with VHL syndrome, mimicking a hypoxic condition due to the inactivation of the HIFa-suppressing pVHL gene. As the SCA cyst epithelium overexpresses a number of hypoxic markers, we sought to determine whether CSPG4 might represent a novel hypoxia-regulated gene. Normal pancreatic ductal epithelial cells (HPDE) and a panel of cancer cell lines were exposed to <1%-oxygen for 3–18 h (acute hypoxia) or 48–72 h (chronic hypoxia), and profiled using qRT-PCR, western blot and FACS-based methods. Indeed, hypoxic conditioning caused an up-regulation of CSPG4 expression; however, this was a restricted phenomenon. First, neither CSPG4 low HPDE cells nor pancreatic cancer cell lines showed significant elevation of CSPG4 expression, although hypoxia greatly increased expression of other hypoxic markers, such as BNIP3, EPO (data not shown), and NIX (Figs. 6A–B). Thus, whatever the reason for CSPG4 silencing in normal ductal cells and the majority of cancer cells, hypoxia was incapable to overcome it. Second, the elevation of CSPG4 mRNA and protein levels in CSPG4 high Panc1 and HS766T cells occurred only upon prolonged (≥48 h) exposure to hypoxia. It should also be noted that hypoxia promoted surface expression of CSPG4 (Fig. 6C), but it did not induce an accumulation of the shed/secreted form (according to ELISA; not shown). Third, CSPG4 transcription was not up-regulated in otherwise hypoxia-sensitive CSPG4 high-primary myofibroblasts/PSCs, the non-transformed cells of non-epithelial origin. This shows that the basal expression alone does not guarantee hypoxic up-regulation. An additional factor is apparently needed.
Figure 6. pCSPG4 as a novel conditional marker of chronic hypoxia in pancreatic tumor cells.
Pancreatic normal (stellate and HPDE) and cancer cells grouped according to the level of constitutive pCSPG4 expression were exposed to hypoxia (0.74% O2) for 3 h and 48 h. QRT-PCR (A), western blot with densitometric quantification using ImageJ software (B) and FACS (C) analyses showed up-regulation of pCSPG4 only in cancer cells with high basal levels of pCSPG4 at 48 h. In contrast, expression of a known hypoxia-sensitive gene, NIX, was strongly up-regulated in all cells.
In combination, these findings indicate that basal expression, mutation/transformation and prolonged oxygen deprivation (physiologic or genetically mimicked) are required for CSPG4 to show hypoxic inductivity. The pathogenic relevance of this induction remains unclear, because, similarly to normoxic cells, the malignant conduct of hypoxic cells (invasiveness and growth) was not affected by siRNA-based knockdowns (not shown).
Discussion
In pancreatic cancer, profiling studies have produced a vast number of putative biomarkers and targets, but very few have accomplished clinical translation. This might be related to the common assumption that an over-expressed molecule is a sign of malignant transformation of an adult ductal cell. Aspects such as extra-pancreatic sources, the systemic character of pancreatic disease, and the still unknown origin of pancreatic malignancies are frequently ignored. The impact of pancreatic disorganization (e.g., parenchymal atrophy, desmoplastic reaction) is rarely taken into account and studies often lack other control groups in addition to the healthy donors.
In our view, this study exemplifies the usefulness of employing a panel of different neoplastic entities as a means of critically evaluating the pathogenic and clinical relevance of the particular molecule. The published records predicted the overexpression and pathogenic relevance of CSPG4 in pancreatic malignancies in line with pro-stromal, pro-angiogenic and pro-malignant activities in other cancers [11]–[15]. Pancreatic expression of pCSPG4 was maintained or elevated in all tested disorders, but most strongly in stroma-poor benign neoplasm SCA. However, levels of sCSPG4 ectodomain shed into patients’ circulation dropped. Selective restoration was observed in advanced or aggressive malignancies, whereby – if at all – pancreatic tumor cells demonstrated overexpression without shedding. Apparently, any pancreatic disease is capable of altering CSPG4 production, whether remote or local, via intrinsic or extrinsic mechanisms. Whereas various cellular sources appear to ensure pCSPG4 synthesis, release of the ectodomain emerges as the main point of interference (block or promotion).
Although the sCSPG4 ‘drop and restoration’ pattern is clearly associated with IPMN and PDAC progression, certain limitations in this study preclude immediate discrimination between direct (cell type-dependent) and indirect (disease-dependent; nodal invasion status) contributions. The sample sizes of the test cohort (n = 83) and validation cohort (n = 221) are relatively small in relation to the variety and genetic heterogeneity of pancreatic diseases, particularly IPMNs [27]–[29], [38]. IPMNs, which may have distinct pathways to cancer progression [30], [31], [39], show an intriguing CSPG4 distinction between non-malignant and malignant entities; only the latter tend to up-regulate both pancreatic and circulating CSPG4 levels. sCSPG4-producing precursors and pericytes are dispersed throughout the body, precluding clear identification of the extra-pancreatic sources to be analyzed. Although we obtained similar results with two different ELISA kits, both assays employed mouse monoclonal antibodies as the capture reagent. The existence of 48 immunologically distinct CSPG4 isoforms and their diversified expression demonstrated recently [19] indicates that a broad panel of antibodies is required to discriminate between total loss and epitope-specific aberration of sCSPG4.
Our observations suggest that pancreatic CSPG4 is not a pathogenic factor promoting pancreatic carcinogenesis. Rather than a pro-malignant bias, a differentially expressed molecule might be a common attribute of a specific lineage and appear as overexpressed once the population starts to expand. It transpires that pancreatic CSPG4 overexpression is a robust feature of a stroma-poor benign adenoma SCA. Its known genetic background (VHL syndrome) allows us to suspect the hypoxic inductivity of CSPG4. The up-regulation of the CSPG4 at 48 h but not 3 h of oxygen reduction indicates a link to chronic hypoxia, thus revealing CSPG4 as a possible target gene of HIF2a; that fits the HIF2a dependency of VHL syndrome [40], [41]. Furthermore, the increased tumor progression seen in a KRAS-driven lung cancer model upon HIF2a depletion (i.e., HIF2a as a tumor suppressor) is in line with the CSPG4’s lack of pro-malignant features [42]. The inability of hypoxia to overcome the CSPG4 negativity of normal adult ductal cells or 78% of cancer cell lines indicates a lineage-restricted pattern of expression, possibly imposed by hypermethylation, as indicated by our experiments with AZA-treated cell lines.
The pancreas is an extremely heterogeneous tissue and the cellular origins of diverse pancreatic tumors are still unclear [43], [44]. Despite the ductal appearance of most exocrine pancreatic malignancies, the transgenic animal models and extensive histological evaluation of biopsies indicate that it might arise from centroacinar cells, terminal ducts, metaplastic acini, or dormant early progenitors [43]–[49]. Although canonical endocrine, acinar and ductal pancreatic lineages originate from the endoderm, the pancreas is infested with stem cells, mesenchymal progenitors and epithelial populations of ambiguous origin [43], [49], [50]. In particular, centroacinar cells and terminal/intercalated ducts are clearly distinct from other cells in the pancreatic ductal system, and demonstrate dramatic expansion in the setting of chronic epithelial injury [51]. Thus, these cells could form the tubular complexes extending among degenerating acini under physiological stress (e.g., hypoxia [49]), and possibly give rise to SCA under genetic mutation. In addition to pancreatic cysts/SCA, VHL syndrome is associated with the development of renal cysts [52], [53] affecting kidney tubular epithelium known to originate from the mesoderm through mesenchymal-to-epithelial transition (MET) [54], [55]. The preferential association of CSPG4 with immature (glial) neural, mesenchymal and epidermal progenitors [7]–[10] suggests that pVHL mutation might affect either a common early precursor (mesendoderm/primitive streak) or a pancreatic epithelial cell of kidney-like mesodermal morphogenesis. Thus, normal pancreata should shelter scarce CSPG4-expressing pluripotent precursors or an obscure differentiated epithelial lineage, both distinct from the adult ductal cells and giving rise to certain neoplasms. CSPG4-expressing PDAC variants are of anaplastic origin (HS766T) or have strong mesenchymal features (Panc1) [56]. These cells might share a common early pancreatic precursor, which might even be the same cell as for SCA but with a different mutation, permitting or facilitating a malignant transformation.
Nonetheless, even knowing which marker should be used, the search for a normal precursor will be a challenge. The shedding of CSPG4, the abundance of circulating isoform and its ability to bind surface-retained extracellular molecules might impede exact identification of the CSPG4-producing precursor. The level of constitutive mRNA expression might be insufficient for in situ detection, although it might become readily detectable once cells are propagating and/or a gene is up-regulated and ‘fixed’ by a hypoxia-mimicking genetic aberration. As an alternative for the derivation of a normal counterpart, one could attempt human islets, shown to contain a distinct population of mesenchymal stem cells able to form CSPG4-positive clusters not of ductal, endothelial, or hematopoietic origin, which however, co-express mesenchymal and pancreatic endocrine markers [10]. Another way might be to establish in vitro systems reproducing the process of pancreatic differentiation [57]. For example, culturing mouse embryonic stem cells on a monolayer of mesonephros-derived cell line M15 replicates a multistep divergence of the ectoderm, mesendoderm and definitive endoderm, all the way to PDX1-expressing progenitors. The latter have been found to mature to all pancreatic lineages (endocrine, exocrine and ductal) upon engraftment under the kidney capsule of SCID mice [58].
While exploring the SCA–hypoxia link, we could not ignore the fact that only 22–57% of SCA cases are associated with VHL syndrome and somatic inactivation of the pVHL gene at chromosome 3p [59], [60]. In 50% of sporadic SCA, tumors have shown allelic loss of a putative tumor suppressor on chromosome 10q [60]. Our screening for hypoxia pathway-related genes identified the HIF1a-inhibitor HIF1AN at the 10q24 locus. Preliminary qRT-PCR measurements revealed a significant reduction of this gene’s mRNA expression in SCA biopsies compared to other pancreatic entities (unpublished observation). Recently, pancreatic deletion of the LKB1/STK11 gene in mice has been shown to result in acinar polarity defects and serous cystic neoplasms [35]. LKB1 mutation also affects hypoxic signaling [36], [37]. In humans, LKB1 loss-of-function mutations have been linked to the development of Peutz-Jeghers syndrome and IPMNs, both high-risk factors for pancreatic cancer [30], [61], [62]. In addition to the implication of candidate mutations for SCA, these data may link the well-established cell/stratum regulatory activity of CSPG4 to cell polarity disorders. It is most likely that (membranous) CSPG4 marks definite epithelial lineage, different from that of an adult pancreatic cell and prone to cyst formation upon ‘pseudo-hypoxic’ CSPG4 overexpression in association with transforming pVHL, HIF1AN and LKB1 mutations.
Altogether, our data indicate that CSPG4 does not contribute to pancreatic carcinogenesis but might be of relevance for cell polarity disorders. CSPG4 emerged as lineage marker, gaining hypoxic sensitivity upon cellular transformation. Surficial retention of CSPG4 on expanding tumor cells improves its standing as a potential therapeutic target [63], [64]. As our study is the first to evaluate circulating sCSPG4, we cannot yet relate our findings to other diseases. The diagnostic relevance of the ‘drop and restoration’ pattern and pancreatic isoforms being clearly distinct (from melanoma-specific antigen), or just unsheddable (in contrast to cervical carcinoma-specific antigens), remains to be explored.
Acknowledgments
We thank E. Soyka, M. Meinhardt and B. Bentzinger (Department of General, Visceral and Transplantation Surgery, University of Heidelberg, Germany) for the excellent technical assistance.
Funding Statement
The work was supported by grants from the Dietmar Hopp Stiftung (http://www.dietmar-hopp-stiftung.de/) to N.A. Giese and from the BMBF to N.A. Giese and A. Bauer (NGFN-01GS08114; http://www.bmbf.de). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hartwig W, Hackert T, Hinz U, Gluth A, Bergmann F, et al. (2011) Pancreatic cancer surgery in the new millennium: better prediction of outcome. Ann Surg 254: 311–319. [DOI] [PubMed] [Google Scholar]
- 2. Erkan M, Michalski CW, Rieder S, Reiser-Erkan C, Abiatari I, et al. (2008) The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol 6: 1155–1161. [DOI] [PubMed] [Google Scholar]
- 3. Ceyhan GO, Bergmann F, Kadihasanoglu M, Altintas B, Demir IE, et al. (2009) Pancreatic neuropathy and neuropathic pain–a comprehensive pathomorphological study of 546 cases. Gastroenterology 136: 177–186 e171. [DOI] [PubMed] [Google Scholar]
- 4. Liu H, Ma Q, Xu Q, Lei J, Li X, et al. (2012) Therapeutic potential of perineural invasion, hypoxia and desmoplasia in pancreatic cancer. Curr Pharm Des 18: 2395–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, et al. (2010) Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science 330: 827–830. [DOI] [PubMed] [Google Scholar]
- 6. Armulik A, Genove G, Betsholtz C (2011) Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev Cell 21: 193–215. [DOI] [PubMed] [Google Scholar]
- 7. Ju PJ, Liu R, Yang HJ, Xia YY, Feng ZW (2012) Clonal analysis for elucidating the lineage potential of embryonic NG2+ cells. Cytotherapy 14: 608–620. [DOI] [PubMed] [Google Scholar]
- 8. Levine JM, Nishiyama A (1996) The NG2 chondroitin sulfate proteoglycan: a multifunctional proteoglycan associated with immature cells. Perspect Dev Neurobiol 3: 245–259. [PubMed] [Google Scholar]
- 9. Legg J, Jensen UB, Broad S, Leigh I, Watt FM (2003) Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development 130: 6049–6063. [DOI] [PubMed] [Google Scholar]
- 10. Carlotti F, Zaldumbide A, Loomans CJ, van Rossenberg E, Engelse M, et al. (2010) Isolated human islets contain a distinct population of mesenchymal stem cells. Islets 2: 164–173. [DOI] [PubMed] [Google Scholar]
- 11. Stallcup WB (2002) The NG2 proteoglycan: past insights and future prospects. J Neurocytol 31: 423–435. [DOI] [PubMed] [Google Scholar]
- 12. Smith FO, Rauch C, Williams DE, March CJ, Arthur D, et al. (1996) The human homologue of rat NG2, a chondroitin sulfate proteoglycan, is not expressed on the cell surface of normal hematopoietic cells but is expressed by acute myeloid leukemia blasts from poor-prognosis patients with abnormalities of chromosome band 11q23. Blood 87: 1123–1133. [PubMed] [Google Scholar]
- 13. Pankova D, Jobe N, Kratochvilova M, Buccione R, Brabek J, et al. (2012) NG2-mediated Rho activation promotes amoeboid invasiveness of cancer cells. Eur J Cell Biol. [DOI] [PubMed] [Google Scholar]
- 14. Svendsen A, Verhoeff JJ, Immervoll H, Brogger JC, Kmiecik J, et al. (2011) Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol 122: 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chekenya M, Krakstad C, Svendsen A, Netland IA, Staalesen V, et al. (2008) The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene 27: 5182–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, et al. (2011) Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One 6: e23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Wang J, Rizvi SM, Jager MJ, Conway RM, et al. (2005) In vitro targeting of NG2 antigen by 213Bi-9.2.27 alpha-immunoconjugate induces cytotoxicity in human uveal melanoma cells. Invest Ophthalmol Vis Sci 46: 4365–4371. [DOI] [PubMed] [Google Scholar]
- 18. Ozerdem U (2006) Targeting pericytes diminishes neovascularization in orthotopic uveal melanoma in nerve/glial antigen 2 proteoglycan knockout mouse. Ophthalmic Res 38: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Girolamo F, Dallatomasina A, Rizzi M, Errede M, Walchli T, et al. (2013) Diversified expression of NG2/CSPG4 isoforms in glioblastoma and human foetal brain identifies pericyte subsets. PLoS One 8: e84883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nishiyama A, Lin XH, Stallcup WB (1995) Generation of truncated forms of the NG2 proteoglycan by cell surface proteolysis. Mol Biol Cell 6: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neesse A, Michl P, Frese KK, Feig C, Cook N, et al. (2011) Stromal biology and therapy in pancreatic cancer. Gut 60: 861–868. [DOI] [PubMed] [Google Scholar]
- 22. Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, et al. (1998) Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 115: 421–432. [DOI] [PubMed] [Google Scholar]
- 23. Erkan M, Kleeff J, Gorbachevski A, Reiser C, Mitkus T, et al. (2007) Periostin creates a tumor-supportive microenvironment in the pancreas by sustaining fibrogenic stellate cell activity. Gastroenterology 132: 1447–1464. [DOI] [PubMed] [Google Scholar]
- 24. Koninger J, Giese NA, di Mola FF, Berberat P, Giese T, et al. (2004) Overexpressed decorin in pancreatic cancer: potential tumor growth inhibition and attenuation of chemotherapeutic action. Clin Cancer Res 10: 4776–4783. [DOI] [PubMed] [Google Scholar]
- 25. Furukawa T, Duguid WP, Rosenberg L, Viallet J, Galloway DA, et al. (1996) Long-term culture and immortalization of epithelial cells from normal adult human pancreatic ducts transfected by the E6E7 gene of human papilloma virus 16. The American journal of pathology 148: 1763–1770. [PMC free article] [PubMed] [Google Scholar]
- 26. Bartel M, Hansch GM, Giese T, Penzel R, Ceyhan G, et al. (2008) Abnormal crosstalk between pancreatic acini and macrophages during the clearance of apoptotic cells in chronic pancreatitis. J Pathol 215: 195–203. [DOI] [PubMed] [Google Scholar]
- 27. Mino-Kenudson M, Fernandez-del Castillo C, Baba Y, Valsangkar NP, Liss AS, et al. (2011) Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut 60: 1712–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tanaka M, Fernandez-del Castillo C, Adsay V, Chari S, Falconi M, et al. (2012) International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology 12: 183–197. [DOI] [PubMed] [Google Scholar]
- 29. Konstantinou F, Syrigos KN, Saif MW (2013) Intraductal papillary mucinous neoplasms of the pancreas (IPMNs): epidemiology, diagnosis and future aspects. JOP 14: 141–144. [DOI] [PubMed] [Google Scholar]
- 30. Amato E, Molin MD, Mafficini A, Yu J, Malleo G, et al. (2014) Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mohri D, Asaoka Y, Ijichi H, Miyabayashi K, Kudo Y, et al. (2012) Different subtypes of intraductal papillary mucinous neoplasm in the pancreas have distinct pathways to pancreatic cancer progression. J Gastroenterol 47: 203–213. [DOI] [PubMed] [Google Scholar]
- 32. Arafat H, Lazar M, Salem K, Chipitsyna G, Gong Q, et al. (2011) Tumor-specific expression and alternative splicing of the COL6A3 gene in pancreatic cancer. Surgery 150: 306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Neumann HP, Dinkel E, Brambs H, Wimmer B, Friedburg H, et al. (1991) Pancreatic lesions in the von Hippel-Lindau syndrome. Gastroenterology 101: 465–471. [DOI] [PubMed] [Google Scholar]
- 34. Mohr VH, Vortmeyer AO, Zhuang Z, Libutti SK, Walther MM, et al. (2000) Histopathology and molecular genetics of multiple cysts and microcystic (serous) adenomas of the pancreas in von Hippel-Lindau patients. Am J Pathol 157: 1615–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hezel AF, Gurumurthy S, Granot Z, Swisa A, Chu GC, et al. (2008) Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol Cell Biol 28: 2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shackelford DB, Vasquez DS, Corbeil J, Wu S, Leblanc M, et al. (2009) mTOR and HIF-1alpha-mediated tumor metabolism in an LKB1 mouse model of Peutz-Jeghers syndrome. Proc Natl Acad Sci U S A 106: 11137–11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, et al. (2014) Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proc Natl Acad Sci U S A 111: 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, et al. (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17: 500–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ideno N, Ohtsuka T, Kono H, Fujiwara K, Oda Y, et al. (2013) Intraductal papillary mucinous neoplasms of the pancreas with distinct pancreatic ductal adenocarcinomas are frequently of gastric subtype. Ann Surg 258: 141–151. [DOI] [PubMed] [Google Scholar]
- 40. Keith B, Johnson RS, Simon MC (2012) HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koh MY, Powis G (2012) Passing the baton: the HIF switch. Trends Biochem Sci 37: 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazumdar J, Hickey MM, Pant DK, Durham AC, Sweet-Cordero A, et al. (2010) HIF-2alpha deletion promotes Kras-driven lung tumor development. Proc Natl Acad Sci U S A 107: 14182–14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kong B, Michalski CW, Erkan M, Friess H, Kleeff J (2011) From tissue turnover to the cell of origin for pancreatic cancer. Nat Rev Gastroenterol Hepatol 8: 467–472. [DOI] [PubMed] [Google Scholar]
- 44. Perez-Mancera PA, Guerra C, Barbacid M, Tuveson DA (2012) What we have learned about pancreatic cancer from mouse models. Gastroenterology 142: 1079–1092. [DOI] [PubMed] [Google Scholar]
- 45. Strobel O, Dor Y, Alsina J, Stirman A, Lauwers G, et al. (2007) In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology 133: 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murtaugh LC, Leach SD (2007) A case of mistaken identity? Nonductal origins of pancreatic “ductal” cancers. Cancer Cell 11: 211–213. [DOI] [PubMed] [Google Scholar]
- 47. Habbe N, Shi G, Meguid RA, Fendrich V, Esni F, et al. (2008) Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A 105: 18913–18918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, et al. (2005) Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell 8: 185–195. [DOI] [PubMed] [Google Scholar]
- 49. Aichler M, Seiler C, Tost M, Siveke J, Mazur PK, et al. (2012) Origin of pancreatic ductal adenocarcinoma from atypical flat lesions: a comparative study in transgenic mice and human tissues. J Pathol 226: 723–734. [DOI] [PubMed] [Google Scholar]
- 50. Esposito I, Seiler C, Bergmann F, Kleeff J, Friess H, et al. (2007) Hypothetical progression model of pancreatic cancer with origin in the centroacinar-acinar compartment. Pancreas 35: 212–217. [DOI] [PubMed] [Google Scholar]
- 51. Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, et al. (2010) Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A 107: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, et al. (2003) von Hippel-Lindau disease. Lancet 361: 2059–2067. [DOI] [PubMed] [Google Scholar]
- 53. Wiesener MS, Maxwell PH, Eckardt KU (2009) Novel insights into the role of the tumor suppressor von Hippel Lindau in cellular differentiation, ciliary biology, and cyst repression. J Mol Med (Berl) 87: 871–877. [DOI] [PubMed] [Google Scholar]
- 54. Stark K, Vainio S, Vassileva G, McMahon AP (1994) Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372: 679–683. [DOI] [PubMed] [Google Scholar]
- 55. Davies JA (1996) Mesenchyme to epithelium transition during development of the mammalian kidney tubule. Acta Anat (Basel) 156: 187–201. [DOI] [PubMed] [Google Scholar]
- 56. Monti P, Marchesi F, Reni M, Mercalli A, Sordi V, et al. (2004) A comprehensive in vitro characterization of pancreatic ductal carcinoma cell line biological behavior and its correlation with the structural and genetic profile. Virchows Arch 445: 236–247. [DOI] [PubMed] [Google Scholar]
- 57. Landsman L, Nijagal A, Whitchurch TJ, Vanderlaan RL, Zimmer WE, et al. (2011) Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biol 9: e1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Higuchi Y, Shiraki N, Kume S (2011) In vitro models of pancreatic differentiation using embryonic stem or induced pluripotent stem cells. Congenit Anom (Kyoto) 51: 21–25. [DOI] [PubMed] [Google Scholar]
- 59. Kim SG, Wu TT, Lee JH, Yun YK, Issa JP, et al. (2003) Comparison of epigenetic and genetic alterations in mucinous cystic neoplasm and serous microcystic adenoma of pancreas. Mod Pathol 16: 1086–1094. [DOI] [PubMed] [Google Scholar]
- 60. Moore PS, Zamboni G, Brighenti A, Lissandrini D, Antonello D, et al. (2001) Molecular characterization of pancreatic serous microcystic adenomas: evidence for a tumor suppressor gene on chromosome 10q. Am J Pathol 158: 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sato N, Rosty C, Jansen M, Fukushima N, Ueki T, et al. (2001) STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. Am J Pathol 159: 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sahin F, Maitra A, Argani P, Sato N, Maehara N, et al. (2003) Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod Pathol 16: 686–691. [DOI] [PubMed] [Google Scholar]
- 63. Poli A, Wang J, Domingues O, Planaguma J, Yan T, et al. (2013) Targeting glioblastoma with NK cells and mAb against NG2/CSPG4 prolongs animal survival. Oncotarget 4: 1527–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geldres C, Savoldo B, Hoyos V, Caruana I, Zhang M, et al. (2014) T Lymphocytes Redirected against the Chondroitin Sulfate Proteoglycan-4 Control the Growth of Multiple Solid Tumors both In Vitro and In Vivo. Clin Cancer Res 20: 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]