Abstract
Population isolates are an important tool in identifying and mapping genes of Mendelian diseases and complex traits. The geographical identification of isolates represents a priority from a genetic and health care standpoint. The purpose of this study is to analyze the spatial distribution of consanguinity by random isonymy (FST) in Argentina and its relationship with the isolates previously identified in the country. FST was estimated from the surname distribution of 22.6 million electors registered for the year 2001 in the 24 provinces, 5 geographical regions, and 510 departments of the country. Statistically significant spatial clustering of FST was determined using the SaTScan V5.1 software. FST exhibited a marked regional and departamental variation, showing the highest values towards the North and West of Argentina. The clusters of high consanguinity by random isonymy followed the same distribution. Recognized Argentinean genetic isolates are mainly localized at the north of the country, in clusters of high inbreeding. Given the availability of listings of surnames in high-capacity storage devices for different countries, estimating FST from them can provide information on inbreeding for all levels of administrative subdivisions, to be used as a demographic variable for the identification of isolates within the country for public health purposes.
Introduction
According to the Hardy–Weinberg law, in an infinitely large population and under panmixia, individuals mate at random and are not biologically related: the population is said to be in equilibrium. However, for social and cultural reasons, among others, human populations tend to shy away from the equilibrium conditions, so that in finite populations all individuals have a certain degree of consanguinity. This contention is supported by genome-wide SNP analyses that have demonstrated the existence of significant portions of homozygosity in the genome above 4 or 5 MB in different populations and in subjects with no record of consanguinity in the last five to ten generations (Bittles 2010; Mac Quillan et al. 2008; Li et al. 2006; Simon Sanchez et al. 2007). Furthermore, in many human populations, the mating between relatives is relatively frequent and stimulated for social, economic, and religious reasons. Overall about 10 % of the world’s population is a result of consanguineous marriages (Bittles 2001; Denic et al. 2011).
Inbreeding coefficients (F) and consanguineous mating types are usually inferred from population surveys (civil registration, inpatients, school surveys, etc.) or pedigree studies. The global prevalence of inbreeding based on the frequency of different types of marriages among relatives, usually up to second cousins, has been estimated from these sources (Bittles and Black 2010) (Global Consanguinity website: www.consang.net). More recently, relying on genomic data, Leutenegger et al. (2011) estimated the inbreeding levels and mating-type proportions in 52 populations from all continents. They found that consanguinity was present in almost all populations with the highest rates of inbreeding in North Africa, the Middle East, and Central South Asia. Leuteneger et al. (2011) concluded that the determination of F based on the genealogies and on genomic data are complementary and their comparison to local or regional levels may provide different results.
Another way to estimate F is provided by the concept of isonymy (I) as defined by Crow and Mange (1965), who observed the relationship between the probability of marriages being isonymyc and consanguineous: the probability of isonymy by descent (I) among all marriages with inbreeding coefficient F would be I = 4F, if all sex combinations of intermediate ancestors of the spouses are equally probable and surnames are monophyletic. This was applied for the estimation of the mean random inbreeding coefficient in the population (FST). The simple formulation of Crow and Mange was repeatedly refined by several authors after 1965 (Yasuda and Morton 1967; Yasuda and Furusho 1971; Yasuda et al. 1974) and applied to study many populations around the world. A synthesis of the literature on isonymy studies is available at the Global Consanguinity website (www.consang.net) and some results obtained in different countries are downloadable from http://www.unife.it/progetti/genetica/pdata.htm. In large heterogeneous societies, the assumptions of the method (monophyletism, absence of illegitimacy or adoption, among others) do not hold and significant differences in the inbreeding levels estimated from isonymy have been observed when mono- and polyphyletic surnames were used (Tay and Yip 1984; Rojas-Alvarado and Garza-Chapa 1994; Garza-Chapa and Rojas-Alvarado 1996). Nevertheless, it has been repeatedly stated that for the indication that drift has occurred, a crude estimate of FST is satisfactory (Yasuda and Morton 1967). Moreover, the relative value of the consanguinity estimates by isonymy is still informative, especially when large sample sizes and the same source of information and methodology are used in an entire country (Relethford 1988).
From the perspective of Medical Genetics, genetic isolates are usually small subpopulations which originated from a small number of individuals with a long history of relative cultural and geographic isolation, high degree of inbreeding, and where a high frequency of individuals affected by rare recessive or infrequent Mendelian diseases can eventually be detected (Boattini et al. 2011). For this reason, population isolates have been used as a tool for mapping and cloning Mendelian disease genes and for studying the genetics of complex traits (Arcos Burgos and Muenke 2002; Peltonen et al. 2000). From this perspective and following the philosophy of community genetics and public health genetics, geographical identification of population isolates represents a priority (Bittles 2001; Ten Kate et al. 2010).
This paper analyzes the spatial distribution of inbreeding by random isonymy (FST) in Argentina and its relationship with genetic isolates previously identified in the country in order to explore the usefulness of this isonymyc parameter of population structure in tracing potential population isolates.
Materials and methods
Calculation and spatial analysis of inbreeding by random isonymy
Information on surnames came from the 2001 electoral register provided by the National Electoral Commission. The names of men and women taken together from the 510 departments, 24 districts or provinces, and 5 geographic regions of Argentina were analyzed. The regions and provinces included are: Northwest (NOA) (provinces of Jujuy, Salta, Santiago del Estero, Catamarca, Tucumán, and La Rioja), Northeast (NEA) (provinces of Formosa, Chaco, Misiones, and Corrientes), Cuyo (provinces of Mendoza, San Juan, and San Luis), Centre (Ciudad Autónoma de Buenos Aires and provinces of Buenos Aires, Entre Ríos, Santa Fe, Cordoba, and La Pampa), and Patagonia (provinces of Neuquén, Santa Cruz, Chubut, Rio Negro, and Ushuaia) were analyzed. The regions used in this analysis correspond to those defined by the Instituto Nacional de Estadísticas y Censos (INDEC) which are based mainly on their geographic distribution, but also on a common history and peopling process of the provinces which they include (Velazquez et al. 2008). Generally speaking, we can say that Argentina population, like others Latin American populations, has a diverse ethnic origin determined by migration from Europe and Africa continents and the consecutive genetic admixture of these allochthonous populations with Native Americans. This process manifested by a heterogeneous degree of genetics admixture according to the region of the country. In the northwest predominate, the Native American component, in the south the European. The African component is minority (Wang et al. 2008; Avena et al. 2012),
Based on the frequency of surnames in each department the expected isonymy was estimated assuming random mating, I = ∑ipi2, where pi is the frequency of surname i in a population, and the summation is over all surnames (Rodríguez Larralde et al. 2011). According to Crow and Mange (1965), FST = (¼) I. This is a rough estimate of the drift that has occurred up to the present in that population (Yasuda and Morton 1967). More details of these calculations are provided elsewhere (Dipierri et al. 2005; Rodríguez Larralde et al. 2011).
Differences in FST between regions were studied with an ANOVA, and its spatial distribution was analyzed as follows: (1) correlating departmental FST values with latitude and longitude of the capital cities of the Departments; (2) using the SaTScan software V5.1 whereby, based on the Poisson distribution, it is possible to detect statistically significant spatial clustering of inbreeding. Due to specific requirements of SaTScan, FST is not a suitable estimator for the software so the B Index (Rodriguez Larralde 1990), defined as the percentage of the population covered by the seven most frequent surnames, was used. This index showed a very high and significant correlation with FST in our analysis (r = 0.98; p < 0.001).
Results
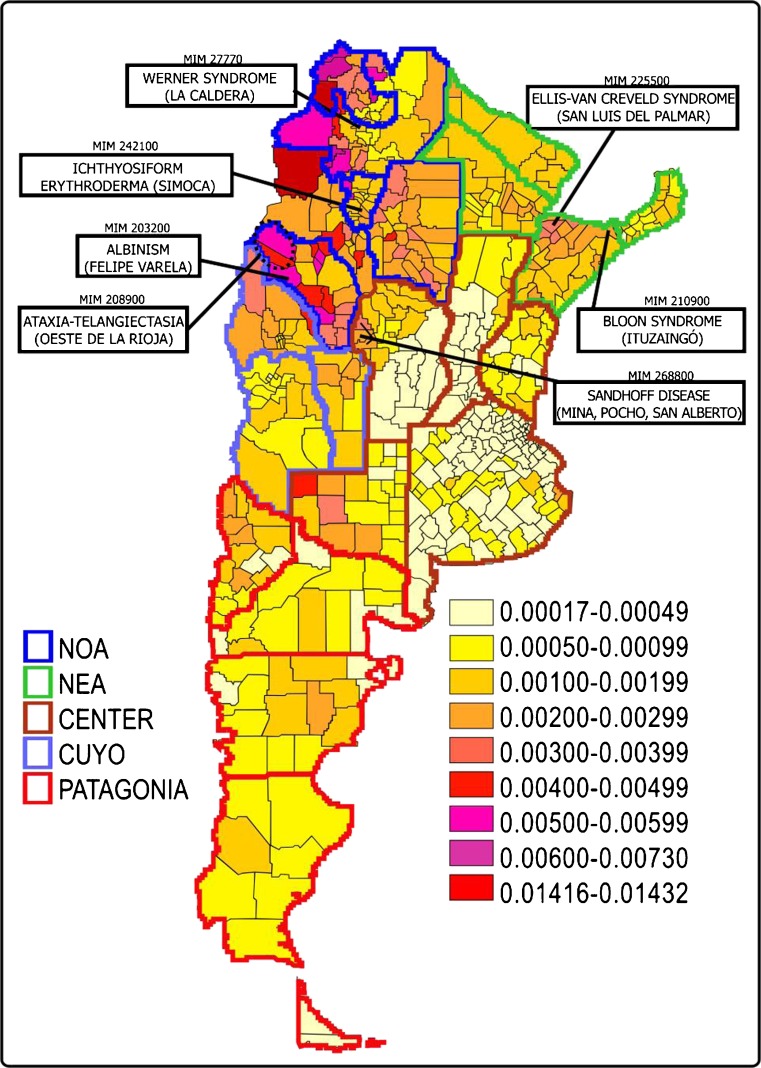
Inbreeding by random isonymy (FST) exhibited a marked variation between departments, the lowest value was 0.00017 (Caseros Department, Santa Fe Province, in the centre of the country) and the highest value was 0.01432 (Susques Department, Jujuy Province, in northern Argentina). Figure 1 presents the distribution of FST at departmental level, where we notice that the highest values were recorded in the northern and western part of the country. FST was highest in the provinces of La Rioja, Corrientes, and Santiago del Estero in the north of the country. It was lowest in the area of Buenos Aires province and in the north-central region of Santa Fe province, both located in the central region of Argentina.
Fig. 1.
F ST distribution by departments and genetic isolated localization
Seventy-three departments with a value of FST ≥ 0.0019 were detected, a magnitude equivalent to the inbreeding value of half third cousins once removed (F = 1/512). Of these, 68 % were located in the northern part of the country. An FST ≥ 0.003 similar to the inbreeding coefficient of half second cousins twice removed (F = 1/256) was observed in 50 departments, of which 90 % also belong to the northern provinces of Argentina: Corrientes, Misiones, Chaco, Formosa, La Rioja, Santiago del Estero, Catamarca, Salta, Tucuman, and Jujuy.
Significant differences in regional FST were detected with an ANOVA, with two outstanding subsets, one consisting of the Centre and Patagonia regions, with lower values of FST and the other, consisting of the regions of NOA, NEA and Cuyo, with higher values. The NOA region was the one with the highest FST values (Table 1).
Table 1.
F ST values: mean, standard deviation (SD), upper and lower limits, by region
| Region | Department numbers | Mean | SD | Lower limit | Upper limit |
|---|---|---|---|---|---|
| Center | 202 | 0.000674 | 0.000580 | 0.000173 | 0.004238 |
| Patagonia | 53 | 0.001475 | 0.000637 | 0.000348 | 0.002860 |
| NOA | 117 | 0.002868 | 0.002138 | 0.000768 | 0.014315 |
| Cuyo | 46 | 0.001475 | 0.000923 | 0.000400 | 0.004850 |
| NEA | 92 | 0.001429 | 0.000725 | 0.000923 | 0.003960 |
NOA Northwestern Argentina, NEA Northeastern Argentina
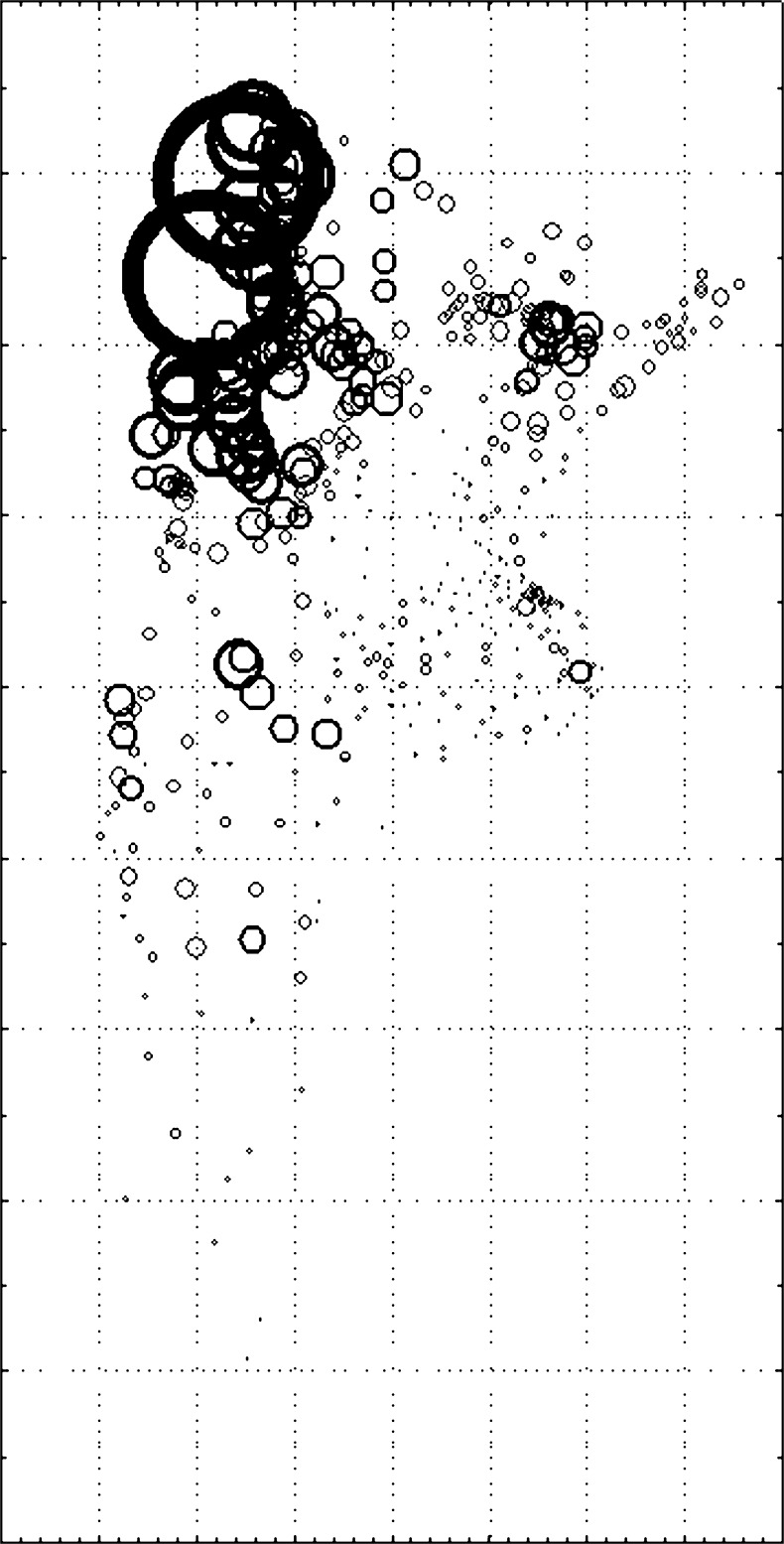
FST correlated negatively with latitude (r = −0.314, p < 0.05) and positively with longitud (r = 0.423, p < 0.05) indicating that the more inbred departments were located towards the north and west of the country (Figs. 1 and 2).
Fig. 2.
Scatterplot of F ST
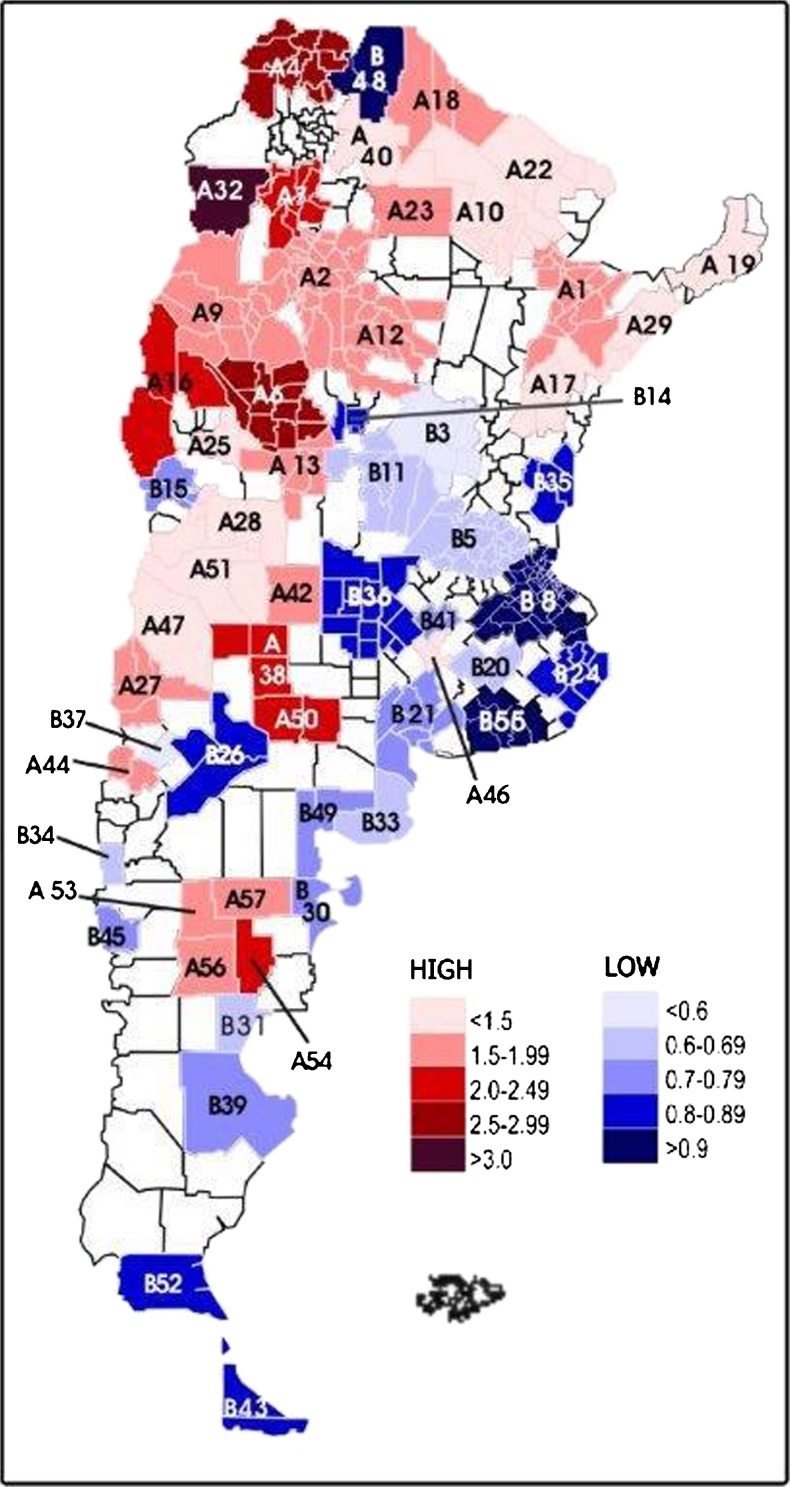
Figure 3 presents the statistically significant FST clusters identified by SaTScan, classified as high (32 clusters) and low (25 clusters) consanguinity, measured by random isonymy. The clusters were formed by a highly variable number of departments ranging from 1 to 31, in the case of high consanguinity. These tend to be located towards the north and west of the country. In the clusters of high inbreeding, the average FST cluster ranged between 0.0142 and 0.0009 and the low inbreeding between 0.0009 and 0.0003. The average FST of the 162 departments that comprise clusters of low inbreeding was 0.00051 ± 0.00023 (with 0.00017 as the lowest value and 0.0022 as the highest) and the average FST of the 196 departments of high inbreeding clusters was 0.002410 ± 0.001784 (with 0.000678 and 0.014315 as lowest and highest values). Of the 510 departments of Argentina, 153 did not belong to any of the groups; in these, the average FST was 0.001111 ± 0.000823 (0.000265 lowest value and 0.005935 the highest).
Fig. 3.
Clusters of high and low consanguinity by isonymy in Argentina
Population isolates in Argentina
In Argentina, some genetic isolates for different autosomal recessive diseases have been detected (De la Fuente et al. 1993; Castilla and Sod 1990; Castilla et al. 1990; Kremer et al. 1985) (Table 2). These were all located on the north and precisely in inbreeding clusters as shown in Fig. 1. Only one of them, found in the Aicuña town, has been extensively studied from the genealogical and molecular viewpoint and these studies showed an 85 % agreement between conventional and molecular genealogies, with mtDNA markers being Amerindian, and Y markers being European (Bailliet et al. 2001).
Table 2.
Population isolates detected in Argentina
| Disease | Acronym | MIM number | Locality | Department (F ST values) | Province | Reference |
|---|---|---|---|---|---|---|
| Oculocutaneous albinism | OCA2 | 203200 | Aicuña | Felipe Varela (0.00534) | La Rioja | Castilla and Adams 1990 |
| Werner syndrome | WRN | 277700 | La Caldera | La Caldera (0.00174) | Salta | De la Fuente et al. 1993 |
| Sandhoff disease | GM2-TYPE II | 268800 | – | Pocho (0.00259) | Córdoba | Kremer et al. 1985 |
| Minas (0.00383) | ||||||
| San Alberto (0.00204) | ||||||
| San Javier (0.00137) | ||||||
| Ataxia-Telangiectasia | AT | 208900 | Aicuña | Felipe Varela (0.00534) | La Rioja | Castilla and Sod 1990 |
| Ellis van Creveld | EVC | 225500 | San Luis del Palmar | San Luis del Palmar (0.00396) | Corrientes | Castilla and Sod 1990 |
| Bloom syndrome | BLM | 210900 | San Luis del Palmar | San Luis del Palmar (0.00396) | Corrientes | Castilla and Sod 1990 |
MIM Mendelian inheritance in man
Discussion
Information on inbreeding was provided in this study by random isonymy at departmental and regional levels. It is worth noting that random isonymy only estimates FST which is not the total inbreeding of the population FIT. Since in Argentina, unlike most of the Spanish speaking countries, only one surname is used, usually that from the father, we miss FIS, the local component of inbreeding, which needs both surnames, paternal and maternal, to be estimated. (Crow and Mange 1965, Wright 1951). Nevertheless a rough estimate of the inbreeding levels of the population is sufficient for the identification of potential genetic isolates.
In this analysis, the lowest administrative subdivision used was the department. In Argentina, these are subdivided into municipalities according to population size. A high FST departmental value suggests that most of its municipalities have high values, and this might be the case for some of the genetic isolates previously detected which include several neighboring departments. On the other hand, a low departmental value does not necessarily imply the absence of genetic isolates within it since localities with higher and lower FST can be present. Then, the analysis of consanguinity through surnames should be deepened to smaller administrative units to have higher chances of identifying potential genetic isolates, but this has not yet been done in Argentina. Although the FST clusters shown in Fig. 3 were identified by SaTScan, using estimator B, as mentioned above, the discussion will be based on FST because of its high significant correlation with B and its frequent use in medical and population genetics. Our analysis shows large variation of FST between regions and an heterogeneous behavior, with its highest values towards the northern and western portion of the country (Figs. 1, 2, and 3).
Studies on the prevalence of consanguinity in Argentina come from different sources and cover different time periods. Orioli et al. (1982) estimated a coefficient and a rate of inbreeding of 0.00011 and 0.3 %, respectively, obtained from the parental consanguinity of malformed children. Castilla et al. (1991) analyzed the frequency of marriages between first cousins in 212,320 civil union certificates dated between 1980 and 1981 across the country. Only 0.37 % of the marriages were between first cousins with an average inbreeding coefficient of 0.00031 for the total sample. Subsequently, Liascovich et al. (2001) confirmed values reported in 1982 by Orioli et al. (1982) in malformed newborns, which were the lowest reported for this kind of source in Latin America. Comparatively, inbreeding values estimated in these studies are similar or lower than those calculated by us with the isonymyc method in the Autonomous City of Buenos Aires and in 23 departments of the provinces of Santa Fe, Cordoba, and Buenos Aires, i.e., in the central part of the country, the most developed and least poor of Argentina. While marriage certificates in Argentina have an item that records whether the civil union is at inbred cousin level, the information provided is relative and unreliable; first, because underreporting has been detected; and secondly, because modern Argentine populations have altered reproductive behavior patterns and determinants of marriage and fertility (Masciadri 2002). The marriage rate began to decline in Argentina in 1950 and currently free unions predominate over legal unions so that unions registered in marriage certificates provide a marriage rate that does not represent the total unions produced in the country. Neither does the relationship between legal unions/free unions present a homogeneous behavior across the country
As stated in the introduction, the inbreeding coefficient of a population can be estimated by genealogies, by the isonymyc method and more recently, through molecular studies. These methods have advantages and disadvantages. The main advantage of the use of surnames is its low cost, it is easy to apply to whole countries or regions, using an appropriate information source and it is little time consuming. Instead, the molecular and genealogy methods, consume much more time, dedication and funding in data collection and subsequent digitalization of information, and problems with participation and collaboration of informants may arise, increasing difficulties for their use with large population groups, countries or departments. Despite these limitations, the genome-wide homozygosity estimation from genomic data has been successfully applied in studies involving a significant number of people (McQuillan et al. 2008; Polasek et al. 2010).
Digital listings of surnames are readily available from various sources of information such as registers of electors, telephone directories, tax payer data bases, civil records, etc. The theory proposed by Crow and Mange (1965) together with the accessibility to a large number of names due to technological developments of digital storage and recording have played an important role, in recent years, in the increase in number of isonymyc studies made at continent (Scapoli et al. 2007), country (Dipierri et al. 2005; Rodriguez Larralde et al. 2000, 2003, 2011) province or state (Rodríguez Larralde and Barrai 1997, 1998), region (Dipierri et al. 2007), and big city (Bronberg et al. 2009) levels, which is why information on inbreeding by random isonymy on populations composed of millions of individuals is available.
The possibility offered by the isonymyc method of describing, in relative terms, consanguinity for different administrative subdivisions would provide an additional highly informative demographic variable that may be related to other biological and sociocultural ones, thus overcoming the mythical conception of inbreeding as “a poor and remote community, a large proportion of whose inhabitants suffer from obscure physical disorders and exhibit obvious symptoms of mental sub normality” (Bittles 1994). According to Pattaro et al. (2007), genetic isolates are not as exceptional, since they are even found in populations with a long history of miscegenation as the Europeans. In many places in Latin America, especially those of small population size, distribution and frequency of surnames mimics onomastics from Macondo, magnificently described by Gabriel García Márquez in “100 Years of Solitude”. According to Castilla and Adams (1996) this would be the onomastic reality of many Latin American rural locations geographically isolated and with high consanguinity. According to Castilla and Adams (1996), in these populations, as in others, rare surnames can serve as genetic markers, identifiers of kinship, and indicators of isolation.
Most of the information on inbreeding in human populations comes from estimations made from unions between closely related relatives (first, double first and first once removed cousins, uncles and nieces, and aunts and nephews), especially in countries with strong cultural tradition to maintain and encourage such unions (North Africa, Central and West Asia and South Asia). Although considerable attention is given to the role of consanguineous marriages as a causal factor of genetic disorders, the potential influence of inbreeding on levels of population homozygosity are still underestimated (Bittles 2005). The isonymyc method can help in reducing this gap, especially in countries with a relatively regular transmission of surnames.
In Argentina, some genetic isolates have been identified in which different autosomal recessive diseases have been detected (De la Fuente et al. 1993; Castilla and Sod 1990; Castilla and Adams et al. 1990, Kremer et al. 1985) (Table 2). Only one of them, found in the town of Aicuña, has been extensively studied from the genealogical and molecular point of view (Bailliet et al. 2001). It is interesting to note that they are all located on the north of the country as shown in Fig. 1, precisely in high inbreeding clusters.
The mapping or cloning of more than 50 genetic disorders in the Finnish, Old Order Amish, Hutterite, Sardinian, and Jewish communities shows that population isolates represent an important tool in studying inherited disorders (Arcos Burgos and Muenke 2002) and in revealing the genetic etiology of common diseases (Pattaro et al. 2007). The geographic location of genetic isolates in Argentina in the clusters with high random consanguinity highlights the usefulness of this approach in identifying isolates with reduced genetic heterogeneity and reduced effective population size. Since the occurrence of rare Mendelian diseases is common in some isolates, their identification, such as selection of candidate populations to be molecularly studied is made (Leutenegger et al. 2011), also has great health care relevance.
The methodology used in this analysis can be applied to other European countries where the isonymy structure of the whole nation has already been published such as Spain (Rodriguez-Larralde et al. 2003), Italy (Barrai et al. 1999) among others and who have nationwide records of rare recessive Mendelian diseases.
Acknowledgments
Ethical standards
This study complies with the current laws of the country in which they were performed.
References
- Arcos Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61:233–247. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- Avena S, Via M, Ziv E, Pérez-Stable EJ, Gignoux CR, Dejean C, Huntsman S, Torres-Mejía G, Dutil J, Matta JL, Beckman K, Burchard EG, Parolin ML, Goicoechea A, Acreche N, Boquet M, Ríos Part Mdel C, Fernández V, Rey J, Stern MC, Carnese RF, Fejerman L. Heterogeneity in genetic admixture across different regions of Argentina. PLoS One. 2012;7(4):e34695. doi: 10.1371/journal.pone.0034695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailliet G, Castilla EE, Adams JP, Orioli IM, Martınez-Marignac VL, Richard SM, Bianchi NO. Correlation between molecular and conventional genealogies in Aicuña: a rural population from Northwestern Argentina. Hum Hered. 2001;51:150–159. doi: 10.1159/000053336. [DOI] [PubMed] [Google Scholar]
- Barrai I, Rodríguez-Larralde A, Mamolini E, Scapoli C. Isonymy and isolation by distance in Italy. Hum Biol. 1999;71:947–961. [PubMed] [Google Scholar]
- Bittles AH. The role and significance of consanguinity as a demographic variable. Popul Dev Rev. 1994;20(39):561–584. doi: 10.2307/2137601. [DOI] [Google Scholar]
- Bittles AH. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60:89–98. doi: 10.1034/j.1399-0004.2001.600201.x. [DOI] [PubMed] [Google Scholar]
- Bittles AH. Endogamy, consanguinity and community disease profiles. Community Genet. 2005;8(1):17–20. doi: 10.1159/000083332. [DOI] [PubMed] [Google Scholar]
- Bittles AH. Time to get real: investigating potential beneficial genetic aspects of consanguinity. Pub Health Genomics. 2010;14(3):169–171. doi: 10.1159/000321772. [DOI] [PubMed] [Google Scholar]
- Bittles AH, Black ML. Consanguinity, human evolution, and complex diseases. PNAS. 2010;107(1):1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boattini A, Griso C, Pettener D. Are ethnic minorities synonymous for genetic isolates? Comparing Walser and Romance populations in the Upper Lys Valley (Western Alps) J Anthropol Sci. 2011;89:161–173. doi: 10.4436/jass.89014. [DOI] [PubMed] [Google Scholar]
- Bronberg RA, Dipierri JE, Alfaro EL, Barrai I, Rodríguez-Larralde A, Castilla EE, Colonna V, Rodríguez-Arroyo G, Bailliet G. Isonymy structure of Buenos Aires city. Hum Biol. 2009;81(4):447–461. doi: 10.3378/027.081.0404. [DOI] [PubMed] [Google Scholar]
- Castilla EE, Adams JP. Migration and genetic structure in an isolated population in Argentina: Aicuña. In: Adams JP, editor. Proceedings of Convergent Questions in Genetics and Demography. Oxford: Oxford University Press; 1990. pp. 45–62. [Google Scholar]
- Castilla E, Adams J. Genealogical information and the structure of rural Latin-American populations: reality and fantasy. Hum Hered. 1996;46(241):a 255. doi: 10.1159/000154361. [DOI] [PubMed] [Google Scholar]
- Castilla EE, Sod R. The surveillance of birth defects in South America: the search of geographic cluster: endemics. Adv Mutagen Res. 1990;2:211–230. doi: 10.1007/978-3-642-75599-6_7. [DOI] [Google Scholar]
- Castilla EE, Gomez MA, Lopez-Camelo JS, Paz JE. Frequency of first-cousin marriages from civil marriage certificates in Argentina. Hum Biol. 1991;63:203–210. [PubMed] [Google Scholar]
- Crow JF, Mange A. Measurements of inbreeding from the frequency of marriages between persons of the same surnames. Eugen Q. 1965;12:199–203. doi: 10.1080/19485565.1965.9987630. [DOI] [PubMed] [Google Scholar]
- De La Fuente S; Tacalitte S; Parada L (1993) Síndrome de Werner: estudios clínicos y genealógicos en varias familias emparentadas del Departamento de La Caldera (Salta). XXIV Congreso Argentino de Genética, pag. 47, Posadas
- Denic S, Naglekerke N, Agarwal MM. On some novel aspects of consanguineous marriages. Pub Health Genomics. 2011;14(3):162–168. doi: 10.1159/000321771. [DOI] [PubMed] [Google Scholar]
- Dipierri JE, Alfaro EL, Scapoli C, Mamolini E, Rodríguez Larralde A, Barrai I. Surnames in Argentina: a population study through isonymy. Am J Phys Anthropol. 2005;128(1):199–299. doi: 10.1002/ajpa.20027. [DOI] [PubMed] [Google Scholar]
- Dipierri JE, Rodríguez-Larralde A, Alfaro EL, Barrai I. Isonymic structure of the Argentine Northwest. Ann Hum Biol. 2007;34(4):498–503. doi: 10.1080/03014460701425427. [DOI] [PubMed] [Google Scholar]
- Garza-Chapa R, Rojas-Alvarado MA (1996). Risk estimation of ABO and Rho(D) incompatibility in persons with mono- and polyphyletic surnames in Monterrey, Mexico. Comparison with other Mexican populations. Arch Med Res 27(2):243–51. [PubMed]
- Kremer RD, Boldini CD, Capra AP, Levstein IM, Bainttein N, Hidalgo PK, Hliba H. Sandhoff disease: 36 cases from Cordoba, Argentina. J Inherit Metab Dis. 1985;8(1):46. doi: 10.1007/BF01805485. [DOI] [PubMed] [Google Scholar]
- Leutenegger AL, Sahbatou M, Gazal S, Cann H, Génin E. Consanguinity around the world: what do the genomic data of the HGDP-CEPH diversity panel tell us? Eur J Hum Genet. 2011;19:583–587. doi: 10.1038/ejhg.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LH, Ho SF, Chen CH, Wei CY, Wong WC, Li LY, Hung SI, Chung WH, Pan WH, Lee MT, Tsai FJ, Chang CF, Wu JY, Chen YT. Long continuous stretches of homozygosity in the human genome. Hum Mutat. 2006;27:1115–1121. doi: 10.1002/humu.20399. [DOI] [PubMed] [Google Scholar]
- Liascovich R, Rittler M, Castilla EE. Consanguinity in South America: demographic aspects. Hum Hered. 2001;51:27–34. doi: 10.1159/000022956. [DOI] [PubMed] [Google Scholar]
- Masciadri V (2002). Tendencias recientes en la constitución y disolución de las uniones en Argentina. Notas de Población, Santiago de Chile: CEPAL 74:53–109.
- McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, Smolej-Narancic N, Janicijevic B, Polašek O, Tenesa A, Macleod AK, Farrington SM, Rudan P, Hayward C, Vitart V, Rudan I, Wild SH, Dunlop MG, Wright AF, Campbell H, Wilson JF. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli IM, Castilla EE, Carvalho WP. Inbreeding in a South-American newborn series. Acta Anthropogenet. 1982;6(1):45–55. [PubMed] [Google Scholar]
- Pattaro C, Marroni F, Riegler A, Mascalzoni D, Pichler I, Volpato CB, Dal Cero U, De Grandi A, Egger C, Eisendle A, Fuchsberger C, Gögele M, Pedrotti S, Pinggera GK, Stefanov SA, Vogl FD, Wiedermann CJ, Meitinger T, Pramstaller PP. The genetic study of three population microisolates in South Tyrol (MICROS): study design and epidemiological perspectives. BMC Med Genet. 2007;5(8):29. doi: 10.1186/1471-2350-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1(3):182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- Polašek O, Hayward C, Bellenguez C, Vitart V, Kolčić I, McQuillan R, Saftić V, Gyllensten U, Wilson JF, Rudan I, Wright AF, Campbell H, Leutenegger AL. Comparative assessment of methods for estimating individual genome-wide homozygosity-by-descent from human genomic data. BMC Genomics. 2010;11:139. doi: 10.1186/1471-2164-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford JH (1988). Estimation of kinship and genetic distance from surnames. Hum Biol 60(3):475–92. [PubMed]
- Rodríguez Larralde A. Distribución de los apellidos y su uso en la estimación de aislamiento y sedentarismo en los municipios del estado Lara, Venezuela. Acta Cient Venez. 1990;41(3):163–170. [Google Scholar]
- Rodríguez-Larralde A, Barrai I. Estructura genético poblacional del Estado Guárico, Venezuela, estimada a través de isonimia. Acta Cient Venez. 1997;48:160–166. [PubMed] [Google Scholar]
- Rodríguez-Larralde A, Barrai I. Estudio genético demográfico del Estado Zulia, Venezuela, a través de isonimia. Acta Cient Venez. 1998;49:134–143. [PubMed] [Google Scholar]
- Rodríguez-Larralde A, Morales J, Barrai I. Surname frequency and the isonymy structure of Venezuela. Am J Hum Biol. 2000;12:352–362. doi: 10.1002/(SICI)1520-6300(200005/06)12:3<352::AID-AJHB5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Larralde A, González-Martín A, Scapoli C, Barrai I. The names in Spain: a study of the isonymy structure of Spain. Am J Phys Anthrop. 2003;121:280–292. doi: 10.1002/ajpa.10209. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Larralde A, Dipierri J, Gomez EA, Scapoli C, Mamolini E, Salvatorelli G, De Lorenzi S, Carrieri A, Barrai I. Surnames in Bolivia: a study of the population of Bolivia through isonymy. Am J Phys Anthropol. 2011;144(2):177–184. doi: 10.1002/ajpa.21379. [DOI] [PubMed] [Google Scholar]
- Rojas-Alvarado MA, Garza-Chapa R (1994). Relationships by isonymy between persons with monophyletic and polyphyletic surnames from the Monterrey metropolitan area, Mexico. Hum Biol 66(6):1021–36. [PubMed]
- Scapoli C, Mamolini E, Carrieri A, Rodriguez-Larralde A, Barrai I. Surnames in Western Europe: a comparison of the subcontinental populations through isonymy. Theor Popul Biol. 2007;71(1):37–48. doi: 10.1016/j.tpb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Scholz S, Fung HC, Matarin M, Hernandez D, Gibbs JR, Britton A, de Vrieze FW, Peckham E, Gwinn-Hardy K, Crawley A, Keen JC, Nash J, Borgaonkar D, Hardy J, Singleton A. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- Tay JSH, Yip WCL. The estimation of inbreeding from isonymy: relationship to the average inbreeding coefficient. Ann Hum Genet. 1984;48:185–194. doi: 10.1111/j.1469-1809.1984.tb01013.x. [DOI] [PubMed] [Google Scholar]
- Ten Kate LP, Al-Gazali L, Anand S, Bittles A, Cassiman JJ, Christianson A, Cornel MC, Hamamy H, Kääriäinen H, Kristoffersson U, Marais D, Penchaszadeh V, Rahman P, Schmidtke J. Community genetics. Its definition 2010. J Community Genet. 2010;1:19–22. doi: 10.1007/s12687-010-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velázquez G (2008) Las regionalizaciones argentinas: evolución de su capacidad de discriminación del bienestar de la población (1943-1992). GEOFOCUS (Revista Internacional de Ciencia y Tecnología de la Información Geográfica) Número 8. http://www.erevistas.csic.es/ficha_articulo.php?url=oai_revista70:251&oai_iden=oai_revista70. Accessed 1 Aug 2013.
- Wang S, Ray N, Rojas W, Parra MV, Bedoya G, Gallo C, Poletti G, Mazzotti G, Hill K, Hurtado AM, Camrena B, Nicolini H, Klitz W, Barrantes R, Molina JA, Freimer NB, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Dipierri JE, Alfaro EL, Bailliet G, Bianchi NO, Llop E, Rothhammer F, Excoffier L, Ruiz-Linares A. Geographic patterns of genome admixture in Latin American mestizos. PLoS One. 2008;4(3):e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetic structure of populations. Ann Eugen (Lond) 1951;15:324–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Yasuda N, Furusho T. Random and nonrandom inbreeding revealed from isonymy study. I. Small cities in Japan. Am J Hum Genet. 1971;23:303–316. [PMC free article] [PubMed] [Google Scholar]
- Yasuda N, Morton NE. Studies on human population structure. In: Crow JF, Neel JV, editors. Third International Congress of Human Genetics. Baltimore: Johns Hopkins Press; 1967. pp. 249–265. [Google Scholar]
- Yasuda N, Cavalli-Sforza LL, Skolnick M, Moroni A. The evolution of surnames: an analysis of their distribution and extinction. Theor Popul Biol. 1974;5:123–142. doi: 10.1016/0040-5809(74)90054-9. [DOI] [PubMed] [Google Scholar]