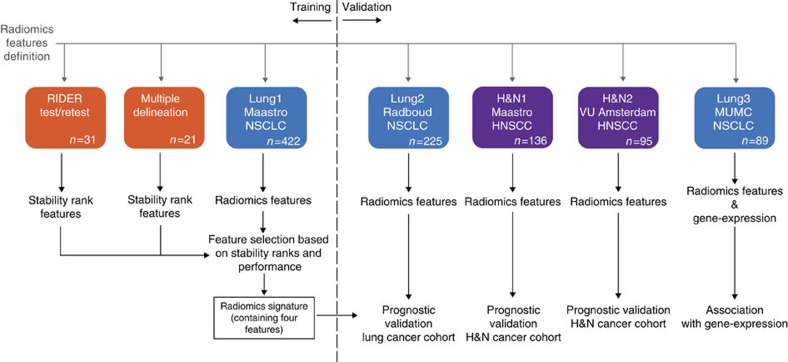
Figure 2. Analysis workflow.
The defined radiomic features algorithms were applied to seven different data sets. Two data sets were used to calculate the feature stability ranks, RIDER test/retest and multiple delineation respectively (both orange). The Lung1 data set, containing data of 422 non-small cell lung cancer (NSCLC) patients, was used as training data set. Lung2 (n=225), H&N1 (n=136) and H&N2 (n=95) were used as validation data sets. The Lung3 data set (n=89) was used for association of the radiomic signature with gene expression profiles. For the multivariate analysis, only one fixed four-feature radiomic signature was tested in the validation data sets.