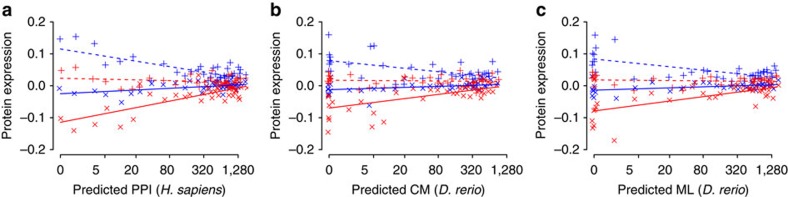
Figure 6. The constraining effect of gene pleiotropy using predicted protein interactions.
(a) Predicted PPI for H. sapiens. GLMM, type II Wald’s F tests with Kenward–Roger df: PPL=1.14E−30, n=960, bootstrap=100%; PEV=5.58E−14, n=960, bootstrap=99%. (b) Predicted CM for D. rerio. PPL=9.11E−10, n=960, bootstrap=98%; PEV=5.39E−05, n=960, bootstrap=85%. (c) Predicted ML for D. rerio. PPL=7.14E−18, n=960, bootstrap=99%; PEV=5.36E−09, n=960, bootstrap=95%. PPL is the significance of the plastic response and PEV is the significance of the evolutionary response in protein expression. PPI, protein–protein interactions; CM, coupling between proteins in the same complex; ML, coupling between proteins in the same metabolic pathway. Predicted PPI, CM and ML numbers were taken from the Funcoup 2.0 database47. CM and ML were used as predictors of PPI for D. rerio. Mild jittering of the points along the x axis was applied to improve plot clarity. Lines are linear regression fits used for visualization. Continuous lines, × 6 °C temperature treatment; dashed lines, +10 °C temperature treatment; blue colour: cold thermal origin; red colour: warm thermal origin.