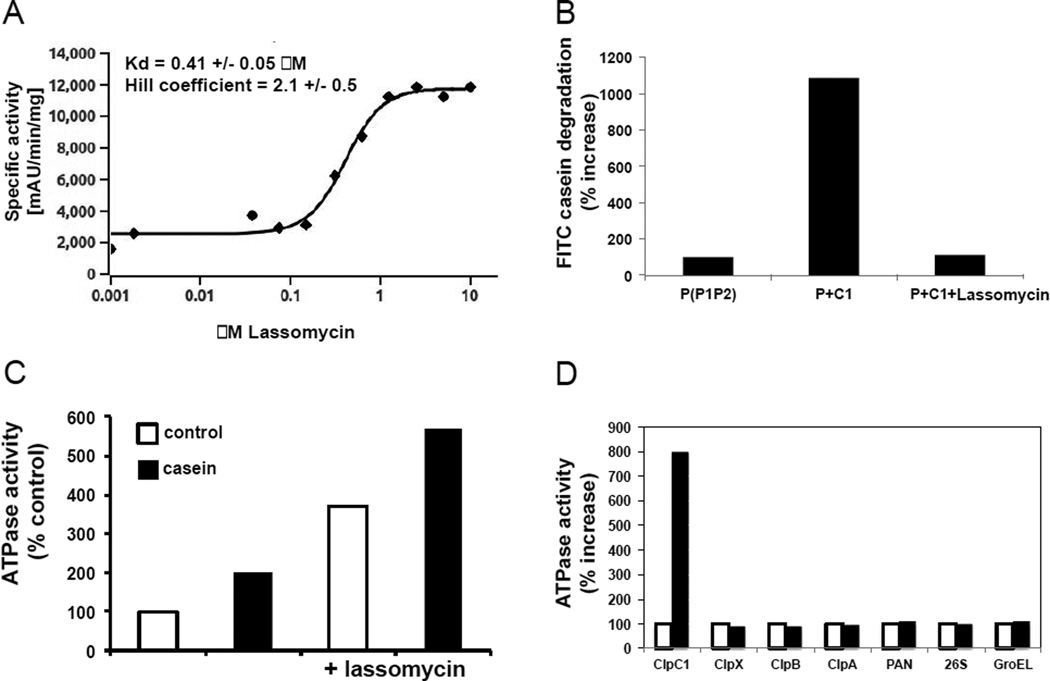
Figure 5. Lassomycin effect on ClpC1 ATPase activity and protein degradation by ClpC1P1P2 complex.
A) Lassomycin stimulates ATPase activity of ClpC1. 32 nM of pure ClpC1 were mixed with 100 μl of the assay buffer (50 mM TrisHCl pH 7.8; 100 mM KCl; 10% glycerol; 1 mM phosphoenolpyruvate; 1 mM NADH; 2 units of pyruvate kinase/lactic dehydrogenase (Sigma); 4 mM MgCl2 and 1 mM ATP) and the ATPase activity of ClpC1 was followed by measuring the coupled oxidation of NADH to NAD+ spectrometrically at 340 nm. Similar results were obtained when the ATPase activity was measured with the Malachite Green method (Geladopoulos et al., 1991). The rate of ATPase activity in the absence of lassomycin was taken as 100%. The apparent Kd and Hill coefficient for lassomycin activation of ClpC1 ATPase were determined using curve fitting with classic Hill-kinetic through a Scaled Levenberg-Marquardt algorithm; tolerance 0.0001. B) ClpC1 does not activate degradation of casein by ClpP1P2 in the presence of lassomycin. ClpP1P2 (100 nM) and ClpC1 (100 nM) were mixed in 80µlofreaction buffer containing 50 mM potassium phosphate (pH 7.6), 100 mM KCl, 8 mM MgCl2, 5% glycerol, 2 mM ATP, 5 mM Z-Leu-Leu. Enzymatic activity was measured fluorometrically using FITC-casein as a substrate in the presence or absence of 10 µM of lassomycin. The rate of degradation of ClpP1P2 was taken as 100%. C) Lassomycin does not interfere with casein binding to ClpC1. ATPase activity of ClpC1 (32nM) was measured as in Fig. 5A in the presence or absence of casein (10 µM) and lassomycin (1 µM). The activity of ClpC1 alone (control) was taken as 100%. D) Stimulation of ClpC1 ATPase activity by lassomycin is highly specific. The activities of purified ATPases from bacteria (M. tuberculosis ClpX; E.coli ClpA, ClpB and GroEL), archaea (PAN) and mouse (26S proteasome) were measured in the presence and absence of lassomycin (10 μM). ATPase activity of each ATPase in the absence of lassomycin was taken as 100%.