Figure 1.
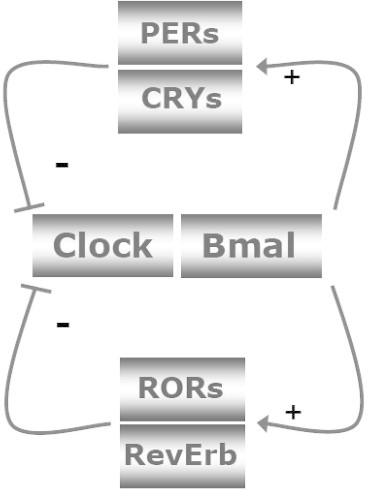
Simplified version of the molecular core clock mechanism. The core loop is formed by Clock:Bmal1 and Period 1–3 (Per1–3) and Cryptochrome 1 and 2 (Cry1–2). The Clock:Bmal1 heterodimer stimulates the transcription of Per1–3 and Cry1–2. Subsequently, Per's and Cry's heterodimerize, translocate to the nucleus, and inhibit Clock:Bmal1 activity. As a consequence, Clock:Bmal1 transcriptional activity drops, which reduces the transcription of Per and Cry genes, thereby activating Clock:Bmal1 again. Additional loops formed by RevErbs and RORs enhance the robustness of the core loop. Post-translational modifications of the clock proteins such as phosphorylation, ubiquitination and sumoylation greatly determine their stability and degradation, which plays a critical role in circadian cycle progression and setting the clock period. For clarity these posttranscriptional regulatory events have been left out of the figure. The feedback loops of the core clock can also regulate widely different phases of genes encoding regulatory components of energy metabolism, by binding to the appropriate promoter elements. On the other hand, changes in energy status have an impact on the molecular clock mechanism. AMPK phoshorylates Cry1 and thereby targets it for degradation. SIRT1 deacetylates, amongst others, Bmal1 and Per2, thereby decreasing the half-life of Per2.
Adapted from Ref. [110].
