Figure 5. Regulation of alternative splicing.
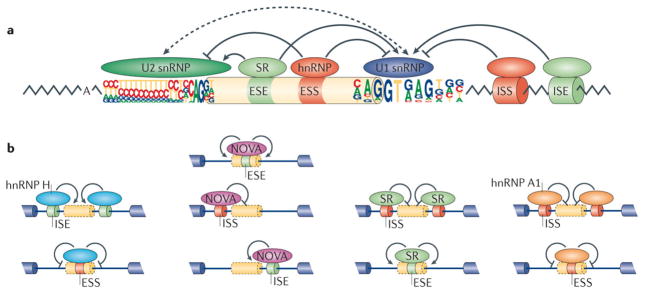
(a) Splice site choice is regulated through cis-acting splicing regulatory elements (SREs) and trans-acting splicing factors. Based on their relative locations and activities, SREs can be classified as exonic or intronic splicing enhancers and silencers (ESEs, ISEs, ESSs or ISSs). These SREs specifically recruit splicing factors to promote or inhibit recognition of nearby splice sites. Common splicing factors include SR proteins that recognize ESEs to promote splicing, as well as various hnRNPs that typically recognize ESSs to inhibit splicing. Both often affect the function of U2 and U1 snRNPs during spliceosomal assembly. (b) The activity of splicing factors and cis-acting SREs is context-dependent. Four well characterized examples are shown from left to right. Oligo-G tracts, recognized by hnRNP H, function as ISEs to promote splicing when they are located inside an intron or as ESSs when located within exons (left). YCAY motifs, recognized by NOVA, act as ESEs when located inside an exon, as ISSs when located in the upstream intron of an alternative exon, or as ISEs when located in the downstream intron. Binding sites for SR proteins or hnRNP A1 also have distinct activities when located at different regions on the pre-mRNA.
