Significance
We report the crystal structure of an intein poised to carry out the rate-limiting step in protein splicing, namely the attack of a conserved Asn side-chain amide on the adjacent backbone amide, leading to resolution of the branched intermediate in the process. The structure reveals that the Asn assumes an unprecedented ready-to-attack conformational state. Guided by this structure, we used protein semisynthesis methods to show that a backbone-to-side-chain hydrogen-bond is critical to position the Asn side-chain for attack and activate it as a nucleophile. This mechanistic insight has general implications for the study of other enzymatic processes involving nucleophilic Asn and Gln residues. The study highlights the power of the combined structural and semisynthesis methods for dissecting protein catalysis.
Keywords: expressed, protein semisynthesis
Abstract
Inteins are autoprocessing domains that cut themselves out of host proteins in a traceless manner. This process, known as protein splicing, involves multiple chemical steps that must be coordinated to ensure fidelity in the process. The committed step in splicing involves attack of a conserved Asn side-chain amide on the adjacent backbone amide, leading to an intein-succinimide product and scission of that peptide bond. This cleavage reaction is stimulated by formation of a branched intermediate in the splicing process. The mechanism by which the Asn side-chain becomes activated as a nucleophile is not understood. Here we solve the crystal structure of an intein trapped in the branched intermediate step in protein splicing. Guided by this structure, we use protein-engineering approaches to show that intein-succinimide formation is critically dependent on a backbone-to-side-chain hydrogen-bond. We propose that this interaction serves to both position the side-chain amide for attack and to activate its nitrogen as a nucleophile. Collectively, these data provide an unprecedented view of an intein poised to carry out the rate-limiting step in protein splicing, shedding light on how a nominally nonnucleophilic group, a primary amide, can become activated in a protein active site.
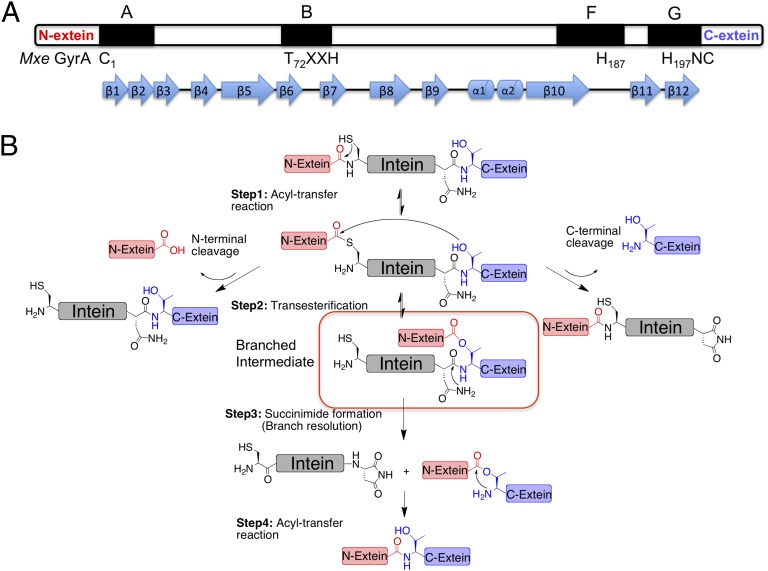
Protein splicing is a posttranslational modification in which an internal domain, termed an intein, excises itself from a host protein with concomitant ligation of the flanking sequences (termed the N- and C-exteins) (1, 2). Inteins are found in unicellular organisms from all domains of life (3) and belong to the HINT (Hedgehog INTein) superfamily of autoprocessing domains (4), which includes the cholesterol ligase domain found in the eponymous hedgehog family of developmental proteins present in all bilaterian animals (5, 6). High-resolution structures of inteins reveal a characteristic horseshoe-like β-sheet fold (7), common to all HINT family members (8), which positions catalytic residues from four conserved sequence blocks (A, B, F, G) in proximity to N- and C-terminal splice junctions (Fig. 1A). This structural information has aided mechanistic studies into the protein splicing process, which we know to be a multistep cascade involving a series of acyl-transfer reactions (Fig. 1B) (1, 2). Although a biological role for protein splicing remains elusive, an ever-deepening understanding of the splicing mechanism has led to the development of a wide range of biotechnology and chemical biology approaches based on engineered inteins (9).
Fig. 1.
Intein-mediated protein splicing. (A) Conserved catalytic blocks (A, B, F, and G) in the intein family; catalytic residues are labeled underneath, β-strands and α-helixes are indicated. (B) Mechanism of protein splicing. The branched intermediate is highlighted. N-terminal and C-terminal cleavage side-reactions are also shown.
The most intriguing chemical step in protein splicing is intein-succinimide formation. This acts as the rate-limiting step in the process and leads to the resolution of the so-called branched (thio)ester intermediate species (Fig. 1B, step 3). This step involves nucleophilic attack of the side-chain primary amide group of a conserved Asn residue (the block G Asn) on the adjacent peptide bond (+1 scissile amide), leading to cleavage of the intein from the C-extein. This is an extremely unusual reaction in proteins (10). Indeed, succinimide formation in proteins more commonly involves attack of a backbone amide on an Asn side-chain, resulting in Asn deamidation (11). A nucleophilic Asn side-chain amide is also associated with N-linked glycosylation, catalyzed by oligosaccharide transferases (12). In this case, two Asn activation mechanisms have been proposed: namely tautomerization of the side-chain amide to a more reactive imidate species (13) and, twisting of the amide C–N bond leading to a more electronegative nitrogen (14). Conceivably, either of these mechanisms could play a role in stimulating intein-succinimide formation. However, the absence of high-resolution structural information on a branched intermediate, from which intein-succinimide formation preferentially occurs, has hindered progress on the issue of how the block G Asn is activated as a nucleophile. It is this structure-activity problem that we set out to address in this study.
Results
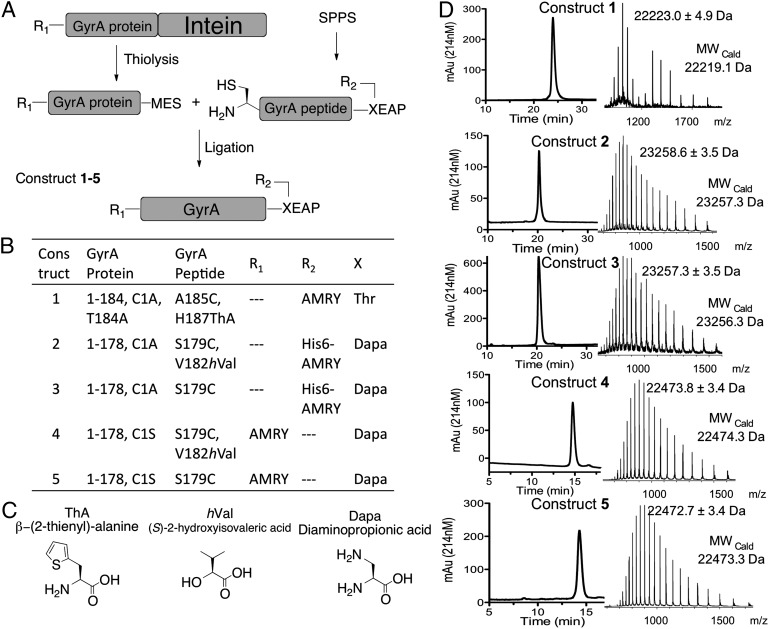
The branched intermediate in protein splicing is an evanescent species and as such presents a formidable challenge for high-resolution structure determination. Previously, we developed a semisynthetic route to the branched intermediate in the Mycobacterium xenopi DNA Gyrase A (Mxe GyrA) intein-splicing reaction (15). Mxe GyrA is the most commonly used intein in protein engineering applications (9). Solution NMR studies on this semisynthetic construct suggest that branched intermediate formation affects the local structure around the +1 scissile amide bond connecting the intein to the C-extein, whereas kinetic studies indicate that intein-succinimide formation is greatly stimulated in the context of this intermediate (15). These studies also suggest an approach to trap the branched structure for high-resolution structure studies, based on mutation of a key histidine residue in the intein, His187 within block F (Fig. 1A); the structure of the Mxe GyrA intein splicing precursor (herein referred to as the linear Mxe GyrA intein structure) reveals this residue is well positioned to act as a general base during succinimide formation (7), and consistent with this role, mutation of this residue selectively blocks this step (15). With this in mind, we used protein semisynthesis to prepare branched construct 1 comprising the Mxe GyrA intein, four native N- and C-extein residues linked through the requisite side-chain ester bond and, critically, the isosteric but noncatalytic unnatural amino acid β-thienyl-alanine (ThA) in place of His187 within the intein (Fig. 2 A–C). This chemically trapped construct was generated in high purity and on a scale suitable for crystallographic analysis (Fig. 2D).
Fig. 2.
Semisynthesis of Mxe GyrA intein constructs. (A) Schematic of the semisynthesis route used in the preparation of protein constructs 1–5. MES refers to a mercaptoethanesulfonate α-thioester. (B) Details of the constructs used in this study. GyrA protein and GyrA peptide refer to the building blocks used in the semisynthesis according to A. R1 and R2 are extein peptide sequences. X is the +1 C-extein residue. (C) Noncoded amino acids used in preparing Mxe GyrA intein constructs. (D) RP-HPLC and ESI-MS analysis of purified constructs 1–5.
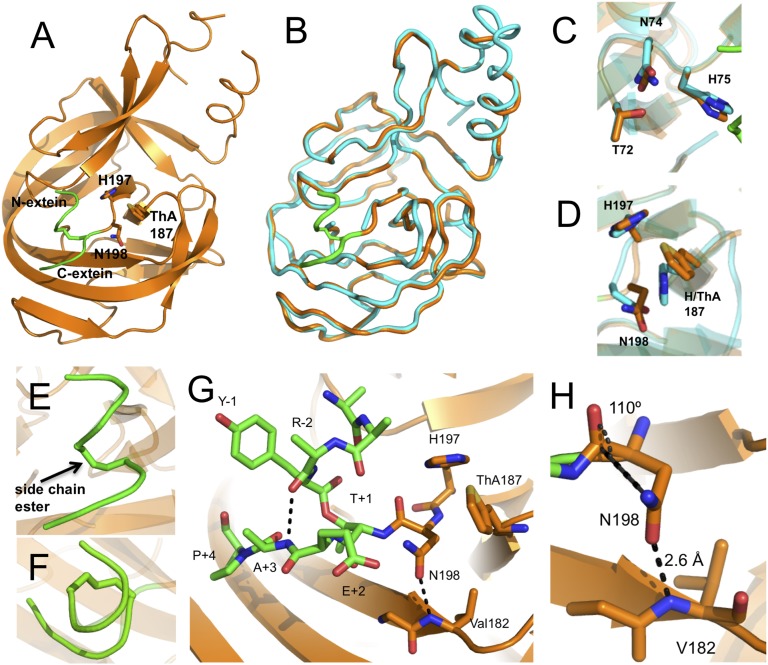
The structure of branched construct 1 was solved by molecular replacement using the linear Mxe GyrA intein structure (PDB ID code 1AM2) (7) as a search model, and refined against 2.79 Å diffraction data (SI Appendix, Table S1). The Mxe GyrA intein branched intermediate structure is nearly identical to its linear splicing precursor, showing the characteristic horseshoe shape with a pseudo C2 symmetry (PDB ID code 4OZ6) (Fig. 3 A and B). The rmsd between the linear and branched Mxe GyrA intein structures is 0.6 Å over the core intein backbone sequence. The side-chains of conserved catalytic residues are also largely unperturbed in the branched structure (Fig. 3 C and D). The greatest deviation is seen in the block F loop (residues 183–188), which assumes a slightly more open conformation in the branched structure compared with the linear (Fig. 3B). This difference is likely because of crystal-packing interactions present in the branched structure that are not observed in the linear Mxe GyrA intein structure (7). The N- and C-extein sequences reside on the surface of the intein and adopt an unusual right-handed corkscrew conformation that includes the branched ester linkage itself (Fig. 3 E and F). This structure appears to be stabilized by a backbone hydrogen bond (H-bond) involving the amide NH of the +3 residue in the C-extein (Ala) and the amide carbonyl of the −2 residue of the N-extein (Arg) (Fig. 3G). In support of this structural inference, this amide NH within the C-extein is necessary for efficient intein-succinimide formation (16), suggesting that a defined branched extein conformation is important for splicing, perhaps by reducing the entropy of this region. Stabilization of this extein structure by a backbone interaction, as opposed to side-chain interactions, is attractive given the known promiscuity of inteins with respect to flanking extein residues, particularly N-extein residues (17, 18). Further to that point, we note that Tyr side-chain at the −1 position in N-extein projects straight into solvent, whereas the side-chains of the remaining three N-extein residues lacked defined electron density and could not be modeled, presumably because of the absence of defined conformations.
Fig. 3.
Crystal structure of the branched intermediate in the Mxe GyrA intein protein splicing reaction. (A) Overall fold of trapped branched intermediate construct 1. The intein secondary structure is rendered in orange and the backbone of the N- and C-extein sequences are in green. Key active-site residues involved in succinimide formation are shown as sticks. The conserved block F His187 residue is replaced by the unnatural amino acid ThA. (B) Backbone overlay of the Mxe GyrA intein structure in the context of the branched intermediate (orange, PDB code 4OZ6) and precursor before protein splicing (cyan, PDB code 1AM2). N- and C-exteins in the branched intermediate structure are colored in green. (C and D) Overlay of conserved block B (C) and F and G (D) active-site residues in the Mxe GyrA branched intermediate (orange) and precursor (cyan) structures. (E and F) Close-up views of the right-handed corkscrew conformation assumed by the ester-linked N-extein and C-exteins in the branched intermediate structure as viewed from the side (E) or from the top (F). (G) Close-up of the Mxe GyrA active site in the branched structure. Exteins are in green sticks and block F and G residues are in orange sticks. Dashed lines indicate apparent H-bonding interactions. (H) Backbone-to-side-chain H-bonding interaction between Val182 and Asn198. The Asn198 side-chain is in a near optimal conformation to attack the +1 scissile amide bond.
The most striking aspect of the branched intermediate structure is the conformation of the block G Asn side-chain (Asn198). This side-chain is well positioned for nucleophilic attack on the +1 scissile amide bond (Fig. 3H and SI Appendix, Fig. S1). Indeed, the angle of approach between the side-chain nitrogen and the backbone amide carbonyl is 110°, which is close to the optimal Bürgi-Dunitz angle for nucleophilic attack on a trigonal unsaturated center (i.e., 107°) (19). This side-chain conformation is not observed in the linear GyrA intein structure (Fig. 3D), nor is it seen in any other intein structures that contain a native block G Asn and a penultimate His (SI Appendix, Fig. S2). The Asn side-chain in the branched structure appears to be held in this conformation by an H-bond between the side-chain oxygen and the backbone NH of Val182 (Fig. 3H). The distance between the donor and acceptor atoms is 2.6 Å, suggesting this is a strong interaction.
To explore the importance of this backbone-to-side-chain H-bond we prepared branched construct 2 containing (S)-2-hydroxyisovaleric acid (hVal) in place of valine at position 182 (Fig. 2). The resulting backbone amide-to-ester substitution removes the H-bond donor and, hence, destroys the interaction between the backbone and the Asn side-chain. Construct 2 was prepared by protein semisynthesis using an analogous route to that used for construct 1, but with a few provisions. First, the ligation site was moved to between residues Tyr178 and Ser179 to allow for backbone engineering at Val182. This ligation strategy required a Ser179Cys mutation to support expressed protein ligation (20). Importantly, this mutation had negligible impact on protein splicing activity of the Mxe GyrA intein (SI Appendix, Fig. S3). Second, the branched peptide fragment contained a diaminopropionic acid (Dapa) residue in place of the +1 Thr residue, and thus the N- and C-exteins are linked by an amide rather than an ester. This circumvented insurmountable synthetic problems (SI Appendix) associated with the preparation of the branched peptide containing both a backbone ester (at Val182) and a side-chain ester (connecting the exteins). Fortunately, we have previously shown that the rate of branched intermediate resolution is unaffected by the substitution of Dapa for the +1 Thr (15). Thus, we could assess the effect of the backbone amide-to-ester substitution at Val182 using this system. Finally, construct 2, as well as control construct 3, which contained a backbone amide at Val182 but retained Dapa at the +1 position, were uniformly labeled with 15N within the recombinant portion of the proteins. Following ligation and purification, constructs 2 and 3 were stored at 4 °C in a phosphate buffer at pH 5.0, conditions known to be compatible with Mxe GyrA intein folding, but that do not allow splicing (15). Hence, we were able to acquire 1H-15N HSQC NMR spectra on semisynthetic constructs 2 and 3. Analysis of these spectra indicated both constructs had assumed the native intein globular fold (SI Appendix, Fig. S4).
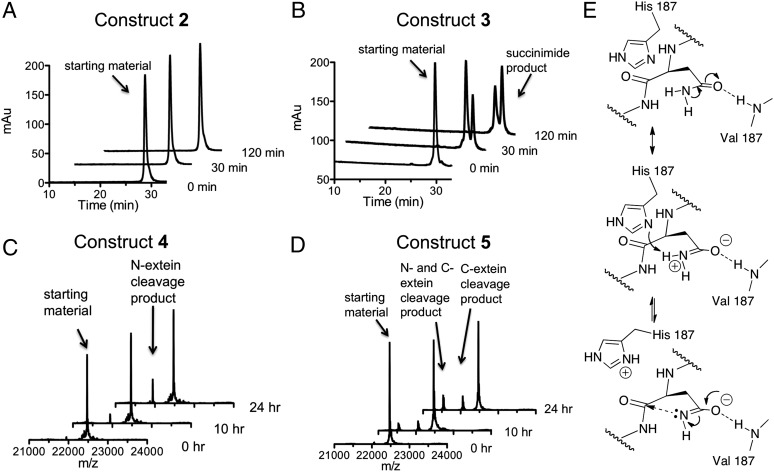
We next compared the rate of branched intermediate resolution in constructs 2 and 3. In each case, the reaction was triggered by raising the pH and temperature of the solution to 7.5 and 25 °C, respectively. Reaction progress was monitored using a combination of reversed-phase HPLC and mass spectrometry (Fig. 4 A and B). Construct 2, which contains an ester bond at Val182, was completely inactive despite being folded. In contrast, construct 3 resolved to the intein-succinimide with a calculated first-order rate constant of 1.0 (± 0.1) × 10−4 s−1 (SI Appendix, Fig. S5) that is in the range expected for this intein (SI Appendix, Fig. S6). This result strongly supports the idea that the H-bond between the amide NH of Val182 and the side-chain of Asn198 is critical for branch intermediate resolution.
Fig. 4.
Backbone engineering of the Mxe GyrA intein. (A and B) Kinetics of succinimide formation for backbone ester-containing branched construct 2 and wild-type branched construct 3 at 25 °C and pH 7.5. Shown are a series of RP-HPLC chromatograms of the reaction mixtures at different time-points. Identities of starting materials and products were determined by ESI-MS. (C and D) Time-course showing the generation of N- and C-terminal cleavage products for backbone ester-containing linear construct 4 and wild-type linear construct 5 at 25 °C and pH 7.5. Shown are a series of deconvoluted ESI-MS chromatographs of the reaction mixtures. Identities of starting materials and products are indicated. (E) Proposed imidate activation mechanism for Asn198 to attack the scissile amide bond: backbone-to-side-chain H-bond stabilizes the resonance form of Asn-198 side-chain amide.
The two main side-reactions in protein splicing are the premature cleavage of the N- and C-exteins before branched intermediate formation (Fig. 1B) (21). Because the latter reaction also involves intein-succinimide formation, we were curious if the backbone-to-side-chain H-bond found to be essential for Asn activation in the branched intermediate is also important in this context. We prepared linear Mxe GyrA splicing construct 4, containing a backbone amide-to-ester substitution at Val182, as well as a Ser in place of Cys1 in the intein (Fig. 2). The latter mutation dramatically attenuates the initiating N-terminal activation step and thus normal protein splicing, but maintains both N- and C-extein cleavage activities (SI Appendix, Fig. S7). As a control, we prepared linear construct 5 that also contains the Cys1-to-Ser mutation but retains the native amide backbone at Val182. Following purification and folding, C-terminal cleavage activity within constructs 4 and 5 was monitored using a mass spectrometry-based assay. Similar to the results on the branched intermediate resolution, succinimide formation was only observed when the backbone amide at Val182 was present (i.e., construct 5 was active, whereas 4 was not) (Fig. 4 C and D). Furthermore, the rate of intein-succinimide formation in construct 5 was significantly slower [2.4 (± 0.2) × 10−6 s−1] (SI Appendix, Fig. S8) than for branched construct 3, consistent with the idea that branched intermediate stimulates this reaction (15). Of note, constructs 4 and 5 both exhibited N-extein cleavage activity (Fig. 4 C and D), which indicates that both proteins adopt an active intein-fold.
Discussion
Our structural studies reveal that the Mxe GyrA intein-fold does not undergo any substantial reorganization to accommodate the branched extein structure. Rather, the linked exteins are perched on top of the intein, where they assume an unusual corkscrew-like structure that has minimal impact on the intein-fold. This arrangement is in keeping with the known promiscuity of inteins with respect to the flanking extein sequences, a property that is integral to their use in protein biotechnology (16, 22). Of the conserved active-site residues in the intein, only two assume an altered conformation in the branched intermediate structure compared with the linear intein precursor, namely the block F His and the block G Asn. The former is likely the result of crystal packing interactions unique to the branched structure and may also relate to the substitution of His for ThA. More interesting is the block G Asn, which appears poised for nucleophilic attack on the +1 scissile backbone amide. The structure identifies a backbone-to-side-chain H-bond as a potential source of this activated conformation. Using protein-engineering methods we were able to confirm that this interaction is essential for intein-succinimide formation in the context of both the branched intermediate and linear precursor. An interesting consequence of this H-bonding interaction would be to stabilize the charged resonance structure of the amide bond, in turn lowering the pKa of the amide –NH2. This process would facilitate tautomerization to the more nucleophilic imidate species via proton shuffling involving the proximal block F histidine residue (Fig. 4D). A similar Asn activation mechanism has been proposed for N-linked glycosylation (13). Thus, this H-bond could play a role in both the positioning and activation of the side-chain amide nucleophile.
Consistent with previous studies (15), we find that intein-succinimide formation is more facile in the context of the branched intermediate. The backbone amide at Val182 is, however, still required for intein-succinimide formation from the linear precursor, implicating the same backbone-to-side-chain H-bond in this context. The observation that Asn198 was not aligned to attack the +1 amide in the linear precursor crystal structure could be a consequence of the truncated construct used in the study (7) (the lack of C-extein residues leaves a negatively charged carboxylate group on Asn198) or could reflect a lower population of the active conformation in the absence of the branch. Solution NMR studies on the Mxe GyrA intein indicate that the +1 scissile amide experiences chemical exchange broadening upon formation of the branched intermediate (15). These data suggests that the +1 scissile amide fluctuates between multiple states, some of which may involve distortion of the bond itself. Based on this observation and those reported herein, we propose that efficient intein-succinimide formation during protein splicing rests on the following features of the system: (i) precise positioning of block G Asn side-chain by the backbone-to-side-chain H-bond identified in the current study, (ii) nucleophilic activation of this Asn by the same H-bond, and, (iii) distortion of the scissile amide bond, which is a unique feature of the branched intermediate.
Methods
Proteins 1–5 used in structural and functional studies were prepared using expressed protein ligation between reactive recombinant and synthetic fragments of the Mxe GyrA intein. The recombinant fragments all contained an α-thioester moiety and were obtained by thiolysis of the corresponding full-length intein fusions, whereas the synthetic building blocks were prepared by Boc or Fmoc solid-phase peptide synthesis. Crystals of protein 1 were obtained by the hanging-drop, vapor-diffusion method and diffracted on a microfocus synchrotron beamline at the Advanced Photon Source. The structure was solved by molecular replacement using the linear Mxe GyrA intein structure as a search model. The coordinates have been deposited in the Protein Data Bank (PDB ID code 4OZ6). Kinetic studies of splicing were performed at pH 7.5 and 25 °C and reaction mixtures at various time-points were analyzed either by SDS/PAGE, RP-HPLC or directly by electrospray ionization (ESI)-MS. Further details are provided in SI Appendix.
Supplementary Material
Acknowledgments
The authors thank the members of the T.W.M. laboratory for valuable discussions; Istvan Pelczer of the Princeton University NMR facility for his generosity; and K. R. Rajashankar and I. Kourinov for support with synchrotron data collection. This work was based in part on research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines, which are supported by Grant P41 GM103403 from the National Institute of General Medical Sciences from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory, was supported by the US Department of Energy under Contract DE-AC02-06CH11357. This work was also supported by the National Institutes of Health Grant GM086868.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.L.S. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4OZ6).
See Commentary on page 8323.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402942111/-/DCSupplemental.
References
- 1.Noren CJ, Wang J, Perler FB. Dissecting the chemistry of protein splicing and its applications. Angew Chem Int Ed Engl. 2000;39(3):450–466. [PubMed] [Google Scholar]
- 2.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 3.Perler FB. InBase: The Intein Database. Nucleic Acids Res. 2002;30(1):383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perler FB. Protein splicing of inteins and hedgehog autoproteolysis: Structure, function, and evolution. Cell. 1998;92(1):1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 5.Porter JA, et al. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374(6520):363–366. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- 6.Ingham PW, McMahon AP. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001;15(23):3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 7.Klabunde T, Sharma S, Telenti A, Jacobs WR, Jr, Sacchettini JC. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat Struct Biol. 1998;5(1):31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 8.Hall TMT, et al. Crystal structure of a Hedgehog autoprocessing domain: Homology between Hedgehog and self-splicing proteins. Cell. 1997;91(1):85–97. doi: 10.1016/s0092-8674(01)80011-8. [DOI] [PubMed] [Google Scholar]
- 9.Vila-Perelló M, Muir TW. Biological applications of protein splicing. Cell. 2010;143(2):191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rawlings ND, Barrett AJ, Bateman A. Asparagine peptide lyases: A seventh catalytic type of proteolytic enzymes. J Biol Chem. 2011;286(44):38321–38328. doi: 10.1074/jbc.M111.260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephenson RC, Clarke S. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J Biol Chem. 1989;264(11):6164–6170. [PubMed] [Google Scholar]
- 12.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 13.Imperiali B, Shannon KL, Unno M, Rickert KW. A mechanistic proposal for asparagine-linked glycosylation. J Am Chem Soc. 1992;114(20):7944–7945. [Google Scholar]
- 14.Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474(7351):350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 15.Frutos S, Goger M, Giovani B, Cowburn D, Muir TW. Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nat Chem Biol. 2010;6(7):527–533. doi: 10.1038/nchembio.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah NH, Eryilmaz E, Cowburn D, Muir TW. Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J Am Chem Soc. 2013;135(15):5839–5847. doi: 10.1021/ja401015p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vila-Perelló M, et al. Streamlined expressed protein ligation using split inteins. J Am Chem Soc. 2013;135(1):286–292. doi: 10.1021/ja309126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27(1):110–114, 116, 118–120. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 19.Burgi HB, Dunitz JD, Lehn JM, Wipff G. Stereochemistry of reaction paths at carbonyl centers. Tetrahedron. 1974;30(12):1563–1572. [Google Scholar]
- 20.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95(12):6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telenti A, et al. The Mycobacterium xenopi GyrA protein splicing element: Characterization of a minimal intein. J Bacteriol. 1997;179(20):6378–6382. doi: 10.1128/jb.179.20.6378-6382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lockless SW, Muir TW. Traceless protein splicing utilizing evolved split inteins. Proc Natl Acad Sci USA. 2009;106(27):10999–11004. doi: 10.1073/pnas.0902964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.