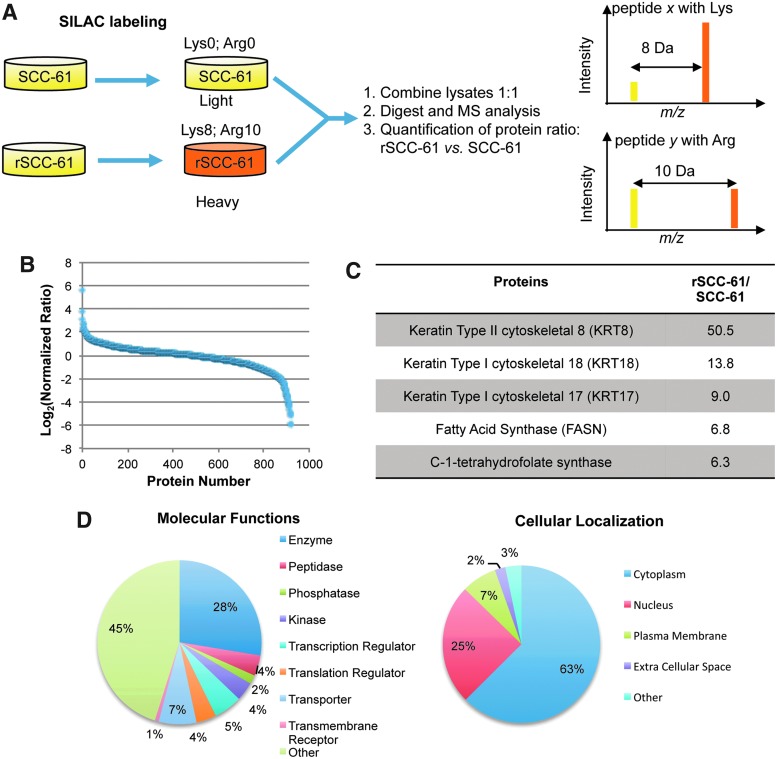
FIG. 2.
Summary of quantitative proteomic analysis of SCC-61 and rSCC-61. (A) Schematic representation of mass spectrometry-based comparative proteomics approach. SCC-61 and rSCC-61 were cultured in media containing light and heavy isotopes of Lys and Arg as described in the “Materials and Methods” section. The normalized mixture of SCC-61 and rSCC-61 proteins lysates was analyzed by mass spectrometry. (B) Protein ratios in rSCC-61 versus SCC-61 (heavy/light). The data were normalized to the median protein ratio. Approximately 30% of the proteins identified had altered protein status in rSCC-61 relative to SCC-61. (C) List of the top upregulated proteins in rSCC-61. The protein ratios generated by the SILAC proteomics analysis were arranged in descending order. An extended list is included in Supplementary Table S3. (D) Cellular localization and functions of the proteins identified in the proteomic analysis. The list of 920 proteins with ratios between 0.017 and 50.5 was input into the IPA. The pie charts represent the classification of the proteins based on their cellular localization or functions. The pie charts corresponding to the entire 965 proteins dataset is included in Supplementary Figure S1. IPA, Ingenuity Pathway Analysis; SILAC, stable isotope labeling with amino acids in cell culture. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars