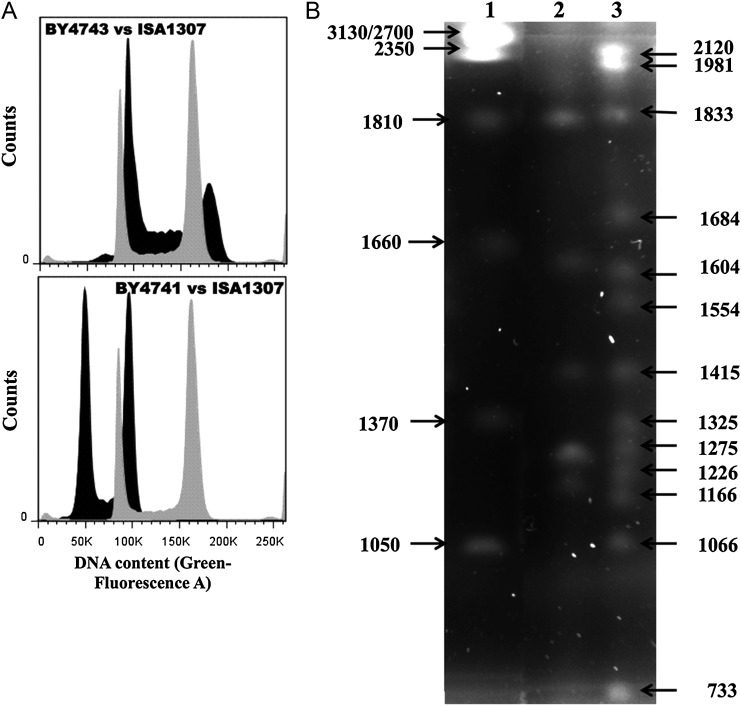
Figure 1.
Estimation of genome size and karyotyping of the ISA1307 strain. (A) Representative cell cycle analysis histogram of S. cerevisiae BY4741 or BY4743 (in black) and ISA1307 (in grey). ISA1307 and S. cerevisiae cells were cultivated in YPD growth medium until stationary phase and then labelled with SYBR Green I to stain genomic DNA. Mean fluorescent intensities (MFI) of G0/G1 peaks of the cell cycle histogram were estimated by flow cytometry. The MFI values obtained for the two S. cerevisiae strains were used to build a calibration curve that was used to calculate the size of the genome of the ISA1307 strain (12.16 Mb for the size of the genome of the haploid strain S. cerevisiae BY4741). (B) Karyotype of the reference strain Z. bailii ATCC58445 (lane 2) and of the ISA1307 strain (lane 3). Total genomic DNA of both yeast species cultivated in YPD growth medium until stationary phase was separated by PFGE. The size of ISA1307 high-molecular-weight chromosomes was estimated based on the high-molecular-weight standard [Hansenula wingei (Bio-Rad)—lane 1], while the size of low-molecular-weight chromosomes was estimated using S. cerevisiae chromosomes' size (not shown).