Figure 8.
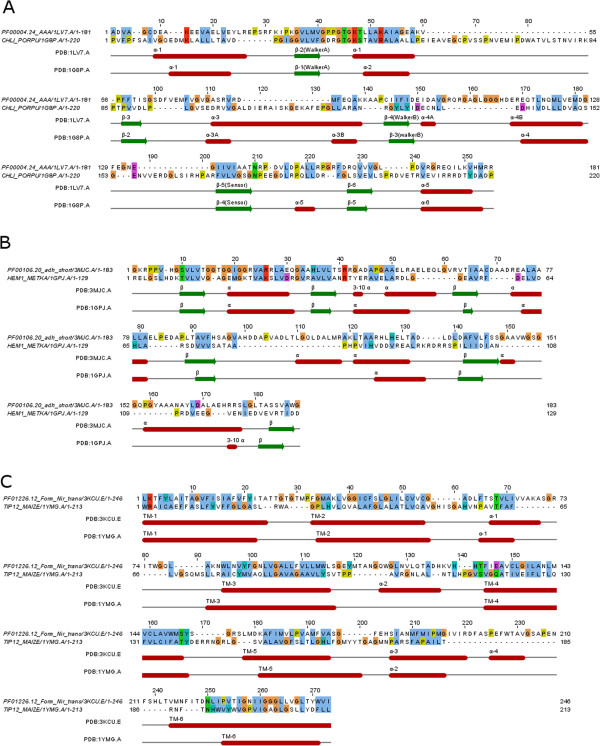
Structural alignments between representative structures of domain model and hit sequence for the 3 true (false-negative) hits. The original E-values of these 3 hits (A. CHLI_PORPU,B. HEM1_METKA, C. TIP12_MAIZE) were insignificant against the Pfam domain models (PF00004.24 AAA, PF00106.20 adh_short, PF01226.12 Form_Nir_trans). However, their structural E-values were nevertheless significant (E < 0.1). Indeed, the structural alignments of representative structures between domain models and hits showed that their RMSD values were between 3.2 to 3.91 and over their full-length sequences. This indicated that the domain model and the associated hit sequences were indeed homologous to each other.
