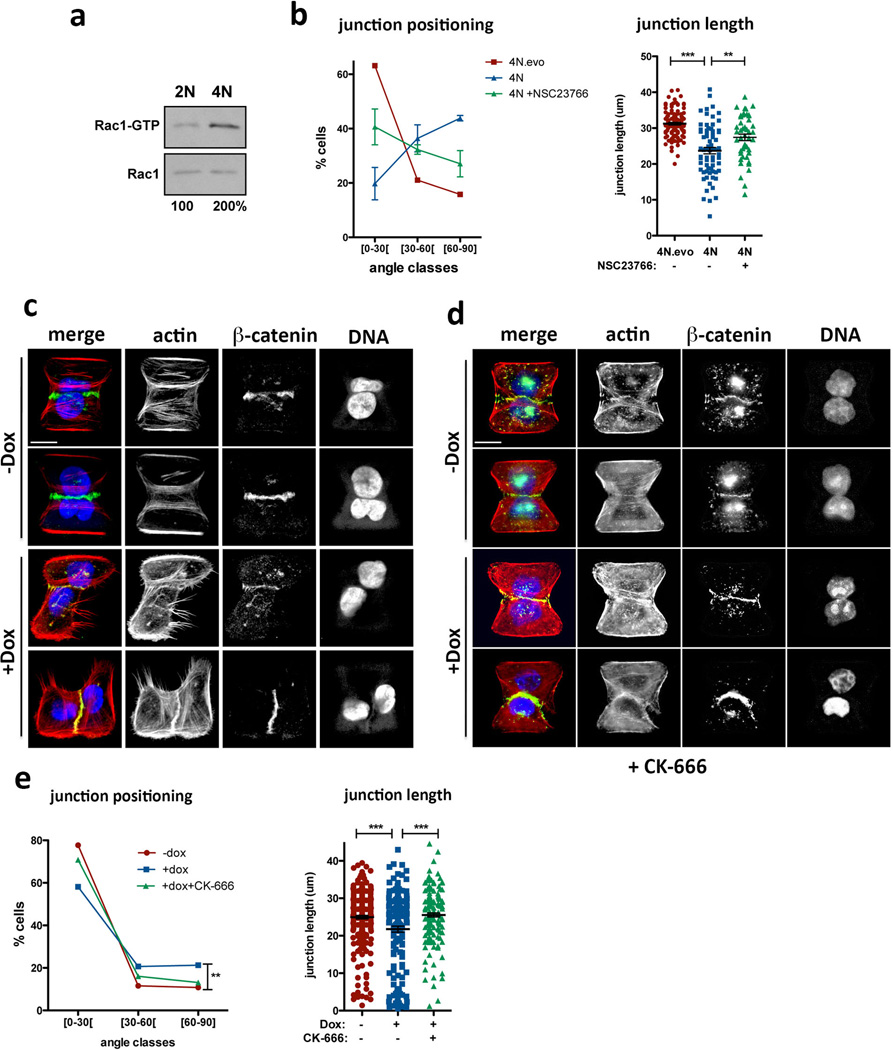
Extended Data Figure 8. Cell-cell adhesion defects caused by centrosome amplification can be observed in tetraploid cells and can be suppressed by Arp2/3 complex inhibition.
a, Western blot from a pull-down experiment to detect GTP-bound Rac1 in tetraploid MCF10A cells. b, Distribution of the cell-cell junction angles (left) and size (right) in the indicated tetraploid cells, with or without treatment with the Rac1 inhibitor, NSC23766. Note that tetraploid cells with extra centrosomes (4N) have a dramatic defect in junction positioning by comparison with tetraploid cells with normal centrosome number (4N.evo). This severe phenotype is only partially rescued by Rac1 inhibition. n4N.evo=106; n4N=70; nNCS23766=47. Error bars represent mean ± SE. c, Examples of cell doublets with (+Dox) or without (−Dox) centrosome amplification on the fibronectin micro-patterns. Cells were stained for F-actin (red), β-catenin (green), DNA (blue). d, Examples of cell doublets with (+Dox) or without (−Dox) centrosome amplification on the fibronectin micro-patterns treated with the Arp2/3 inhibitor (CK-666). Cells were stained for F-actin (red), β-catenin (green), DNA (blue). e, Distribution of the junction angle and quantification of the junction size in cells with extra centrosomes treated with 50 µM of Arp2/3 inhibitor (CK-666) for 6hrs. Cells were analyzed 48hrs after Dox treatment. n−Dox=251; n+Dox=160; nCK666=168. Error bars represent mean ± SE. All the p-values were derived from unpaired two-tailed t-test (***, p<0.0005; **, p=0.005). Scale bar: 10µm.