Abstract
Crystal polymorphs of glucose isomerase were examined to characterize the properties and to quantify the energetics of protein crystal growth. Transitions of polymorph stability were measured in poly(ethylene glycol)/NaCl solutions, and one transition point was singled out for more detailed quantitative analysis. Single crystal x-ray diffraction was used to confirm space groups and identify complementary crystal structures. Crystal polymorph stability was found to depend on the NaCl concentration, with stability transitions requiring > 1 M NaCl combined with a low concentration of PEG. Both salting-in and salting-out behavior was observed and was found to differ for the two polymorphs. For NaCl concentrations above the observed polymorph transition, the increase in solubility of the less stable polymorph together with an increase in the osmotic second virial coefficient suggests that changes in protein hydration upon addition of salt may explain the experimental trends. A combination of atomistic and continuum models was employed to dissect this behavior. Molecular dynamics simulations of the solvent environment were interpreted using quasi-chemical theory to understand changes in protein hydration as a function of NaCl concentration. The results suggest that protein surface hydration and Na+ binding may introduce steric barriers to contact formation, resulting in polymorph selection.
Introduction
The ability of proteins to crystallize is derived from a small fraction of the protein surface that contributes to crystal contacts, and because of the number and types of interactions that are possible, multiple crystal polymorphs are often encountered. Polymorphic crystals that form under similar conditions allow for the identification of crystal structures with potentially similar free energies of crystallization. Hence, polymorphic crystals offer an opportunity to discern the interactions responsible for crystallization as well as those required to maintain a stable polymorph.
The identification and refinement of methods to select for a preferred crystal polymorph is likely no more important than in the case where protein crystals are to be used as therapeutic protein delivery vectors.1–4 As an example, Pechenov et al.2 studied the dissolution behavior of a model protein, α-amylase, with two different crystal polymorphs suspended in gel formulations. The difference in dissolution rates between the crystal polymorphs was significant with one morphology showing up to 80 % dissolution compared to ca. 30 % for the other over the same time.
Several physicochemical methods have been employed to identify solution conditions suitable for protein crystal growth, and as a result many proteins have been shown to exhibit crystal polymorphism. Crystal polymorphs of aprotinin (BPTI) have been extensively studied5–7 and their stability was found to depend on pH, salt and temperature. One polymorphic crystal of BPTI displays a retrograde solubility profile with temperature.7 Polymorphs of α-amylase were selected using crystallization at different ionic strengths2 or temperatures.8 High concentrations of (NH4)2SO4 (> 1.5 M) were required to nucleate different polymorphic crystals of glucose isomerase (GI),9 while urate oxidase was shown to produce different crystal polymorphs as a function of PEG concentration,10 which was attributed to the modification of long-range interactions. The enzyme ribonuclease A has been successfully crystallized in a significant variety of space groups using widely different solution additives (e.g., salts, alcohols and polymers).11–13
An even more interesting system is that of L-asparaginase, where the polymorphs retain the same space group but differ in their crystal lattice contacts, specifically due to Ca2+ binding.14 The presence of ions in solution during crystallization is normally associated with screening of electrostatic interactions, while higher ionic strengths are normally used to precipitate proteins via salting-out. Ion binding, however, has been observed to affect the crystal lattice for lysozyme,15 urate oxidase,16 thaumatin17,18 leucine-binding protein19 and histidine-binding protein.20 More detailed studies concerning the interactions of ions and protein surfaces have shown the preference of Na+ ions for carboxylate groups,21 although the additional effect of protein hydration due to the presence of the ion has not received as much scrutiny. Lund et al.22 have shown that the level of hydration of a specific ion does correlate to its preference for protein surfaces. Surface-force measurements have provided additional evidence of repulsive hydration forces due to hydrated bound cations on mica surfaces that can be scaled relative to the cation dehydration energy.23–26
GI is one of the relatively small number of proteins that are purified by crystallization on a large scale, in quantities of tons/year.9 GI is a large tetrameric protein with a molecular weight of 172 kDa and a pI of ~4, estimated from the amino acid sequence. Although the protein is unstable at lower pH and denatures below pH 5, it is quite robust at higher pH, being most active in the pH range 7 to 9 and at temperatures of 60 to 80°C, depending on the source of the enzyme.27 Interestingly, in (NH4)2SO4 solutions, GI is least soluble at pH 7,9 quite far from the pI.
Because of its prolific use at industrial scales the crystallization of GI from numerous organisms has been well-studied. For example, the crystallization of GI from Arthrobacter B3728 in ammonium sulfate solutions was observed to produce three polymorphs.28 Use of the additive thymol was found to determine which polymorph resulted at high (NH4)2SO4 concentrations (> 1.5 M).29 GI from Streptomyces rubiginosus was observed to crystallize into at least two polymorphs with (NH4)2SO4aline9 and with poly(ethylene glycol) (PEG) and NaCl.30
In this work we sought to identify experimentally solution conditions that show a preference for a specific crystal form of GI, while identifying a mechanism for polymorph selection. In most reports of polymorphic protein crystals a mechanism of polymorphic crystal selection is often assumed or inferred, but rarely explored quantitatively. We report the experimental identification of transitions between two polymorphic crystal forms of GI, which are observed to have distinctly different gross morphologies, referred to here as polyhedral and rectangular. Confirmation of polymorphism was provided by single-crystal x-ray diffraction and normalized osmotic second virial coefficients (b2) were measured by self-interaction chromatography (SIC) to probe the net protein interactions around an experimentally identified polymorphic crystal transition. Using crystal structures obtained from the Protein Data Bank (PDB),31 a multi-scale molecular mechanics method was employed to estimate the interaction free energies of the crystal contacts in order to determine the dominant interactions of each polymorph. Additionally, molecular dynamics simulations were performed on fragments of the protein, with and without NaCl, to account for local protein surface hydration and potential ion binding, facilitating a discussion of the implications of hydration and ion-binding for crystallization and polymorphism.
Experimental
Materials
GI from Streptomyces rubiginosus was obtained from Macrocrystal Oy (Finland) as a crystal slurry and was used as received. Xylitol (X-3375), poly(ethylene glycol) 10 kDa (PEG; 81280), N-hydroxysuccinimide (NHS; 130672) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC; E6383) were from Sigma. PEG 8 kDa (P156-500), hydrochloric acid (HCl; A144-212), sodium hydroxide (NaOH; SS410-4) and sodium chloride (NaCl; S271-500) were from Fisher Scientific. Toluene (61095-1000) and 4-morpholinoethanesulfonic acid (MES; 172595000) were from Acros. The Micro BCA protein assay kit (23235) and 10k MWCO Slide-A-Lyzer® dialysis cassettes (66830) were from Pierce. Toyopearl AF-Amino-650M resin particles (08002) were purchased from Tosoh Biosep. An AP-Mini 5 x 100 mm (WAT064-02) chromatography column was from Waters. Paraffin oil (HR3-421) and 72-well microbatch plates (HR3-087) were from Hampton Research.
Solutions
Solutions were prepared using distilled deionized water further purified using a Milli-Q Plus system (Millipore). All solutions were buffered with 10 mM MES at pH 6.5, unless otherwise noted. Stock solutions containing either 5 M NaCl or 40% (w/v) PEG were used for the preparation of subsequent samples. The solution pH was adjusted using 6 M HCl or NaOH. All buffers were filtered through 0.22 μm sterile bottle-top filters (Pall), NaCl stock solutions were filtered through 0.22 μm sterile syringe filters and stock PEG solutions were filtered through 0.8 μm sterile syringe filters (Millipore).
GI was received as a crystal slurry in a solution of ammonium sulfate and magnesium sulfate. A slurry sample was first dialyzed against water until the solution became clear. Dialysis was continued against 10 mM MES pH 6.5 buffer for approximately 24 hours. The protein solution was filtered through a 0.22 μm sterile syringe filter (Millipore) and then concentrated using either a 10 or a 30 kDa Amicon Ultra4 centrifugal filter (Millipore). Protein concentrations were measured by absorbance using a Perkin-Elmer Lambda 4B UV-visible spectrophotometer at a wavelength of 280 nm using an extinction coefficient of 1.0 cm2 mg–1.30
Simultaneous Crystal Dissolution Observations
Polyhedral and rectangular crystals to be used for transfer and subsequent study were grown by homogeneous crystallization methods in microbatch plates at defined conditions that all included NaCl and 10 kDa PEG. Microbatch plates for studying crystal dissolution were prepared at specific solution conditions of PEG, NaCl and GI, typically in 10 μL total volumes. Initial protein concentration ranges were from 1–25 mg ml–1. Observable dissolution of a crystal type indicated a lack of stability of that polymorph under the specific solution conditions. Prior to the introduction of crystals, each well was checked visually to ensure that no crystals or aggregates were present. Crystals of each polymorph were transferred from their respective growth wells to a well of known composition using Mitegen microtools (Mitegen). Prior to release of crystals, each tip was blotted with a Kimwipe® to minimize transfer of growth solutions in addition to that of micro-crystals. All wells were covered with a layer of paraffin oil to prevent evaporation and consequent concentration changes. Crystal dissolution and/or growth was observed optically using an Olympus BH2 upright microscope and images were taken using a Nikon Coolpix 8700 digital camera. Solution ranges were 2–6% (w/v) PEG 8 or kDa 10 kDa with 1–2 M NaCl. Visualization of crystal growth was not a requirement to identify a transition in stability. Each condition was examined at least in duplicate.
Equilibrium Solubility Measurements
Crystals of each type, polyhedral and rectangular, were prepared separately by seeding. Polyhedral crystals were grown from a supersaturated solution (>50 mg ml–1) in 0.17 M MgSO4 in 10 mM bis-tris, pH 6.9, at 22 ± 2°C.9 Rectangular crystals were grown from a supersaturated solution (>50 mg ml–1) in 1.75 M (NH4)2SO4 in 10 mM bis-tris, pH 7, at 22 ± 2°C.9 Solutions of known NaCl and PEG concentrations in which the solubility was to be measured were prepared without protein. To minimize transfer of growth solutions, crystals of a single morphology were transferred to a 500 μL centrifuge tube and centrifuged at 14000 g for 10 min. The supernatant was removed and the crystals were centrifuged again. Any remaining supernatant was removed. The crystals were then washed with 500 μL of solution at the NaCl and PEG concentrations of interest, mixed for 5 min and centrifuged again. The supernatant was discarded and replaced with a fresh aliquot of the same composition, in which dissolution was allowed to occur. The soluble protein concentration was monitored regularly using absorbance at 280 nm. Equilibrium was normally achieved within 1 week, but samples were also checked after two weeks for consistency. The crystal morphology was verified using optical microscopy.
X-ray Diffraction
X-ray diffraction data sets were collected for a single crystal of each crystal form on a Rigaku RU300 rotating anode generator with a R-AXIS IV image plate area detector. Crystals were soaked in a solution of the mother liquor with xylitol added as a cryo-protectant. In each case the crystal was flash-frozen in liquid nitrogen. The polyhedral crystals were grown in 0.4 M NaCl, 4% (w/v) PEG 10 kDa at 4°C. A xylitol concentration of 60% of the saturation value was required to obviate development of an ice ring. The rectangular crystals were grown in 1.75 M NaCl, 6% (w/v) PEG 10 kDa at 22 ± 2°C. A xylitol concentration of 50% of the saturation value was required for an ice ring to be absent. Each crystal was set for 1° oscillation along the ∞ axis and exposed for 10 minutes per angle. All data were processed using HKL2000.32 For the polyhedral crystals 36 frames were analyzed, whereas 90 frames were analyzed for the rectangular crystals.
Self-interaction Chromatography
Immobilization
Immobilization of GI on Toyopearl AF-Amino-650M resin particles was performed using EDC/NHS chemistry.33,34 Approximately 2 mL of resin was washed extensively with water. A solution of 150 mM EDC and 7 mM NHS was prepared in 10 mM MES buffer, pH 6.5. The resin was resuspended in the EDC/NHS solution with 30 mg of GI to a total volume of 13 mL and left on a 360°C rotisserie overnight. Using more than 30 mg of GI per immobilization experiment resulted in insoluble aggregates. In order to achieve a practically useful immobilized-GI concentration another immobilization step was required, therefore the resin was separated from the supernatant and a fresh solution of EDC/NHS with another 30 mg of GI was added and left for 10 hrs. Following the reaction the resin was washed with 500 mL aliquots of water, 10 mM MES at pH 6.5, 1 M NaCl in 10 mM MES at pH 6.5 and finally 10 mM MES at pH 6.5. An identical preparation without GI was used as the blank. The immobilized-GI concentration was measured using the Micro BCA protein assay. A final surface coverage of ~16% (18.9 ± 0.5 mg GI ml–1 settled resin) was determined and used for all measurements.
Chromatographic measurements
The resin was packed in a Waters AP-Mini 5 × 100 mm column at a flow rate of 1.4 ml min–1. All measurements were made at 22 ± 2°C using an ÄKTA Purifier 10 with detection by absorbance at 280 nm. All runs followed essentially the same steps as described previously.33–36 Samples were injected in 40 μL volumes with a GI concentration of ~6 mg ml–1. A base flow rate of 0.1 ml min–1 was used, while samples run in the presence of PEG were at reduced flow rates to account for the change in effective protein diffusivity due to the increase in solution viscosity.33 The retention volume was determined using the first moment of the peak rather than the peak maximum,35,36 evaluated using a Matlab routine developed in-house. From the retention volume the retention factor was calculated using36
| (1) |
where Vr is the retention volume in the GI-immobilized column at a specific solution composition and V0 is the column dead volume. The surface coverage and injection concentration were not observed to alter the results appreciably for coverages ranging from 7% to 38% and GI injection concentrations of 2 mg mL–1 to 12 mg mL–1.
Virial coefficient calculations
B22 is related to the retention factor (k′) by36
| (2) |
where BHS represents the hard-sphere or excluded-volume contribution, ρs is the amount of protein immobilized per unit area, and ϕ is the phase ratio.37BHS was estimated as 16πr3/3, where r is the equivalent protein radius. Small-angle x-ray scattering experiments38 yielded a radius of 3.3 nm, and this value was used here.
B22 as calculated in eqn. 2 is usually reported in units of mol ml g–2, which tends to result in small absolute values for larger proteins. Therefore a normalized virial coefficient has been proposed as a more appropriate means of comparison among proteins.34,39 The normalized virial coefficient, referred to here as b2, is calculated relative to BHS, so that for SIC
| (3) |
Static Light Scattering
Because SIC is a relatively new method for virial coefficient measurements, values determined by this method are often compared for validation to those obtained by static light scattering (SLS). Here b2 was measured by SLS at 90° using a Brookhaven light scattering apparatus with a BI9000AT correlator and a Lexal (Fremont, CA) model 95 Ar-ion laser (λ = 488 nm). All measurements were performed at 25 ± 0.1 °C, controlled by an external circulating bath. Absolute Rayleigh ratios (R90) were calculated based on a Rayleigh value (Rt) for toluene of 30.7 × 10–6 cm–140 by41
| (4) |
where the measured scattered intensities of the protein (Ip), the protein-free solution (Ib) and toluene (It) are all at 90°.
Rayleigh ratios were used to determine b2 by the method of Zimm,42
| (5) |
where cp is the protein concentration in g ml–1, MW is the protein molecular weight and R90 is the Rayleigh ratio at 90°. K is an optical constant related to the properties of the solution and the instrument, and is given by43
| (6) |
where n0 is the refractive index of the solvent, λ is the wavelength of the laser light in a vacuum, NA is Avogadro's number and dn/dc is the refractive index increment of the protein solution. The dn/dc value for GI was determined to be 0.1424 ± 0.0004 ml g–1 using a differential refractometer (C. N. Wood, RF-600) at 535 nm.
Modeling and Simulation
Identification of Crystal Contacts
Identification of the residues involved in crystal contacts can provide useful information to help identify a molecular mechanism for polymorph selection. Using crystal structures from the PDB31 (accession codes 2GLK44 and 1OAD45) that x-ray diffraction data show correspond to the polyhedral and rectangular crystals respectively, specific residues participating in the crystal contacts were identified. The asymmetric units provided within the PDB files were used to generate the native tetramer and contact residues were identified as participating in either direct residue-residue interactions or in water-mediated interactions. Direct residue-residue interactions were defined as those with heavy-atom center-to-center distances of 4 Å or less. These account for short-range interactions between hydrogen bonding groups and between non-polar groups. Water-mediated interactions required each participating protein atom to be a hydrogen bond donor or acceptor, i.e., oxygen or nitrogen atoms, and to be within 3.5 Å of an intervening water molecule. No orientational constraints were applied. Contacts identified in this manner are in agreement with those from the alternative PISA46 program. The contact area in each crystal form was determined using the Molecular Surface Package.47 Individual contact areas were determined as one-half the difference in the total solvent-accessible area of two molecules in contact relative to that of the two independent molecules.
Crystal Contact Interaction Free Energy
The interaction free energy of crystal contacts was estimated using a multi-scale molecular mechanics approach that has been extensively documented previously.48,49 The approach considers two types of interactions, which in earlier work49 were parameterized relative to experimental protein-protein binding free energies and so are assumed here to yield reasonable estimates of the interaction free energy. Short-ranged, non-electrostatic interactions are calculated using a hybrid Lifshitz-Hamaker (LH)/Lennard-Jones (LJ) model together with an atomically detailed description of the protein. Briefly, for heavy atom pairs separated by less than 6 Å the scaled LJ model (with parameters obtained from the OPLS-UA force field50) is used, while for atom pairs beyond this threshold the LH model is used, with a protein-water-protein Hamaker constant of 3.1 kT.51 As in earlier studies,48,52 for electrostatic interactions the protein was treated as a spherical dielectric cavity of radius 3.3 nm (here obtained from SAXS38) with angular placement of charge as found in the protein. Charge as a function of pH is determined using intrinsic pK values.53,54 a Charges due to divalent cations contribute an additional +8e, so that the net charge is –54e at pH 6.5. The Poisson and linearized Poisson-Boltzmann equations were solved in their respective domains using the boundary element method55 with a protein dielectric constant of 4 and the solvent dielectric constant of 80, with the Debye screening length determined by the salt concentration of interest.
Molecular Dynamics
The effects of water on crystal contact interaction free energies were examined by incorporating water molecules into the molecular mechanics calculations above in either of two ways. In the first, all crystallographic water was assumed to be part of the protein and considered to interact with adjacent protein molecules in the same way as protein atoms, using the methods described in the previous section. In the second, molecular dynamics (MD) simulations were performed to identify those water molecules that are strongly associated with the protein surface; these simulations were interpreted using quasi-chemical theory,41,52,56 where strongly associated water molecules can be considered to be an intrinsic part of the protein.56
Each GI molecule comprises over 12000 atoms, and we are interested in the interaction between two GI molecules, so the associated computational cost for an explicit treatment of the entire protein (together with the solvent) is prohibitive. However, the observation that proximal protein-water radial distribution functions depend mainly on the chemical identity of the protein atom in question and are less sensitive to the broader environment57,58 suggests that we can safely consider only protein fragments that constitute the protein-protein interface. Each fragment was chosen such that within the molecular mechanics model, the interaction free energy was within 1 kT of that obtained using all atoms. Protein atoms in each fragment were fixed, consistent with the rigid-body description in the molecular mechanics models. Water occupancy times are expected to be an order of magnitude shorter than large-scale protein conformational changes, and hence the rigid body approximation is acceptable for our purposes. This approximation also constitutes a focus on the specific protein conformation represented by the crystal structure as opposed to an averaging over the ensemble of possible protein conformations.
MD simulations were performed in the NPT ensemble using NAMD 2.659 for two different systems: (1) the fragments were simulated separately (infinite separation case) and (2) two fragments from separate molecules were simulated as a complex, as in the crystal contact. Fragments were protonated and solvated using VMD60 with the CHARMM27 force field.61 Water was represented using the TIP3P62 model and the geometry was constrained by the SETTLE algorithm.63 Na+ and Cl– ions were added using the Autoionize plugin in VMD. The non-bonded LJ interactions were gradually switched to zero between 9.5 Å and 10 Å; no appreciable changes in the number and location of bound water molecules were found in test simulations with the LJ interactions switched to zero between 10 Å and 12 Å . Electrostatic interactions were calculated using the particle-mesh Ewald method64 with a grid spacing of ~1 Å. An integration time-step of 2 fs was used, and the simulations were performed at a constant temperature of 298 K through the application of the Langevin dynamics method to all non-hydrogen atoms, with a damping coefficient of 1 psps–1. The pressure was maintained at 1 bar using the Nose-Hoover Langevin piston65 with a period of 200 fs and a decay time of 100 fs. Each run consisted of 50000 steps of steepest-descent minimization followed by 10 ps of heating to 298 K. A 0.5 ns equilibration step was followed by a final production run of 4 ns. Configurations were saved every 0.2 ps for analysis.
Method to locate bound water molecules
The location of bound water molecules in proximity to the protein surface was determined using a 1 Å grid over the protein heavy atoms.41,52 The grid was restricted to the first hydration layer, i.e., less than 3.5 Å from the protein surface. The local density of water at each grid site (ρ) was determined and related to the excess chemical potential (Δμex) of the water through41,52
| (7) |
where ρb is estimated as the real bulk water density (0.0333 Å–3). Each grid site with η ≥ 2 was identified as a location of a single bound water molecule,41,52 i.e., the water molecule is considered bound if its excess free energy is 2 kT lower than the bulk value.
Results and Discussion
Identification of Stability Transition
The more stable of the two crystal polymorphs of GI at a given solution condition was determined using simultaneous crystal dissolution experiments, an example of which is shown in Figure 1. The microscopic changes in the crystals clearly reflect differences in the relative stability of the two different crystal habits, observed as polyhedrals or rectangles. Under these solution conditions the polyhedral crystals are observed to dissolve into the bulk solution while the rectangular crystal edges sharpen, indicating that the rectangular crystals are the more stable. A strength of this technique is that a small number of crystals and a small sample size are adequate to observe differences in stability. Offsetting this is that the polyhedral crystals in this example do not fully dissolve into the bulk solution. It is likely that without mixing the local GI concentration is too high to allow sufficient dissolution, and as a result remnants of the original crystals remain. The polyhedral remnants may also include small nucleated rectangular crystals.
Figure 1.

Example of polyhedral and rectangular crystals in 4% PEG 10 kDa, 1.5 M NaCl, 10 mM MES, 5 mg mL–1 GI pH 6.5 at 22 ± 2°C. Polyhedral crystals dissolve while the edges of rectangular crystals sharpen, indicating rectangle stability. Scale bar represents 250 μm.
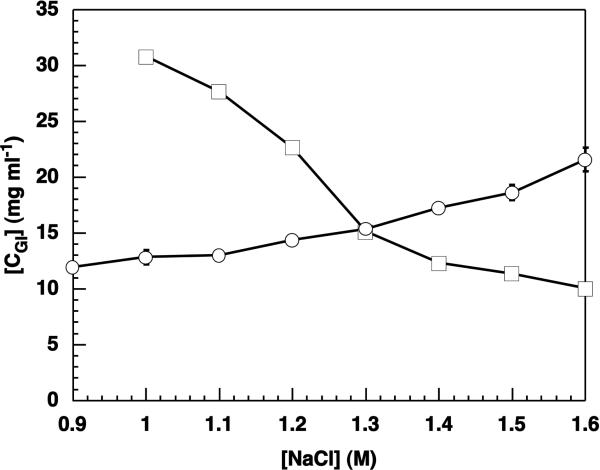
Solution conditions at which there are crystal stability transitions from polyhedral to rectangular as determined by simultaneous crystal dissolution are given in Table 1. At the resolution of the data obtained, the transitions are controlled solely by the NaCl concentration, and are largely insensitive to the PEG concentration. However, all of the conditions at which transitions were measured included PEG; crystallization did not occur, nor did the crystals remain stable, in the absence of PEG, up to protein concentrations of ~ 250 mg ml–1. The presence of PEG therefore appears to be necessary to induce or maintain crystallization when NaCl is used but does not seem to control polymorph selection. In contrast, for the urate oxidase10 system, the PEG concentration was observed to control polymorph selection. The observation of polymorph selection caused by different influences highlights the sensitivity of protein-protein interactions as they apply to polymorphic crystals.
Table 1.
Polymorph transition ranges as a function of PEG molecular weight, PEG concentration and NaCl concentration at 22 ± 2 °C. [NaCl] identifies the range below which the polyhedral crystals are more stable and above which the rectangular crystals are more stable.
| PEG MW (kDa) | % (w/v) PEG | [NaCl] (M) |
|---|---|---|
| 8 | 2 | 1.1 - 1.2 |
| 8 | 4 | 1.2 - 1.3 |
| 10 | 2 | 1.1 - 1.2 |
| 10 | 4 | 1.3 - 1.4 |
Previous studies of the crystallization of GI using (NH4)2SO4 without PEG required (NH4)2SO4 concentrations ranging from 1.5–1.9 M before rectangular crystals were observed.9 In each case the requirement of a high salt concentration indicates that long-range electrostatics are not significant in the crystal contacts and that the interactions in crystals are likely dominated by short-range interactions, such as van der Waals, salt bridging and hydration interactions. Polymorph selection of rectangular crystals grown in the presence of (NH4)2SO4 also required temperatures ≥25°C. Solutions of NaCl and PEG are also temperature-sensitive in that when solutions at conditions for which rectangular crystals are more stable at room temperature were kept at 4°C, only polyhedral crystals nucleated.
As a final method of verification of polymorphism between the polyhedral and rectangular crystals, single-crystal x-ray diffraction was performed. Table 2 shows the space groups and unit cell dimensions of each crystal form along with unit cell parameters for two GI structures deposited in the PDB.31 The unit cell dimensions for each crystal type are sufficiently close to those for the solved structures to allow the structures here to be assumed to be the same as the respective deposited structures. It is also noteworthy that the growth units in each case are single tetramers and not larger oligomers with different salts and organic additives,44,45 similar to what has been reported for BPTI polymorphs.66
Table 2.
Unit-cell properties for the polyhedral and rectangular polymorphs and two crystal structures of the same space groups. 2GLK and 1OAD are PDB accession codes.
Polymorphic crystals are normally in different space groups by definition, although the same space groups have been observed with differences in the intermolecular contacts.14 The fact that the two crystal types exhibit differences in solubility under identical solution conditions (Figure 1) is further evidence that these crystals contain different intermolecular contacts. Vivares et al.10 used similar reasoning with urate oxidase polymorphs. In their case, the equilibrium temperature was used to probe crystal stability, allowing identification of true polymorphs as opposed to habit changes (macroscopic differences due to accelerated growth along a particular axis). Ultimately, the concentration of PEG 8 kDa or the temperature was determined to control polymorph selection and this was attributed to changes in long-range interactions modulating protein orientation.
Equilibrium Solubility
Equilibrium solubility measurements were performed by independent crystal dissolution in the vicinity of an identified transition in stability between the two crystal forms. The range considered was 4% (w/v) PEG 10 kDa, 1.3-1.4 M NaCl, since this was the first transition identified by simultaneous dissolution (Table 1). Figure 2 shows the equilibrium solubility of both the rectangular and polyhedral polymorphs as a function of NaCl concentration. Excellent agreement is obtained with the previous simultaneous dissolution experiments in locating the transition at approximately 1.3 M NaCl. The solubility of polyhedral crystals is observed to increase with increasing NaCl concentration, most notably at NaCl concentrations above the stability transition from polyhedral to rectangular. This increase can be seen as a salting-in effect, although such effects are normally observed at low salt concentrations. In contrast, the rectangular crystals exhibit the normal salting-out behavior observed for most proteins. While Figure 2 spans NaCl concentrations from 0.9 to 1.6 M NaCl, measurement of rectangular crystal solubility was not possible at 0.9 M NaCl because of the nucleation of polyhedral crystals in less than one week. By extrapolating the rectangular crystal solubility line to 0.9 M NaCl the solubility is estimated to be roughly three times that of the polyhedral crystals, which is often cited as a rule-of-thumb for the concentration necessary to observe homogeneous nucleation.67,68 At the opposite end of the NaCl concentration range, at 1.6 M NaCl rectangular crystals were often observed nucleating homogeneously within the polyhedral samples within about two weeks, which may account for the slightly larger error bars.
Figure 2.
Equilibrium solubility lines of rectangular (□) and polyhedral (○) polymorphs measured independently. The crystal type with the lower solubility at each [NaCl] is the more stable form. Error bars are from duplicate measurements. Lines are meant to guide the eye.
The observation that there is a high NaCl requirement for polymorph transition suggests that multiple mechanisms may contribute to the interactions within each crystal type. Again, long-range electrostatics are not expected to be a factor in the transition, but short-range van der Waals interactions may be influential, while ion binding and changes in protein hydration may also play a role.
Normalized Osmotic Second Virial Coefficients
To probe the protein-protein interactions more directly, normalized osmotic second virial coefficients (b2) were measured by SIC. Virial coefficients were also measured without PEG by SLS and SIC to validate the SIC method. The presence of PEG when performing SLS is a complicating feature in that it requires consideration of not only protein-protein interactions but also protein-PEG interactions69 and scattering by PEG. SIC proves to be advantageous in this respect since PEG-protein interactions may influence b2, but no changes in methodology or analysis are required for the experimental conditions with and without PEG.
At NaCl concentrations ≥0.3 M the results from SLS and SIC show adequate agreement. However, at lower NaCl concentrations a deviation was observed for SIC values compared to SLS, suggesting that the SIC values are less reliable at these conditions due to possible protein-column interactions, although experiments with the blank column did not indicate that this was the case. This may be caused by the Donnan contribution.41,52 Despite the uncertainties regarding the results at low salt, the measurements that are relevant here are at high NaCl concentrations, where there is much better agreement between the two complementary methods.
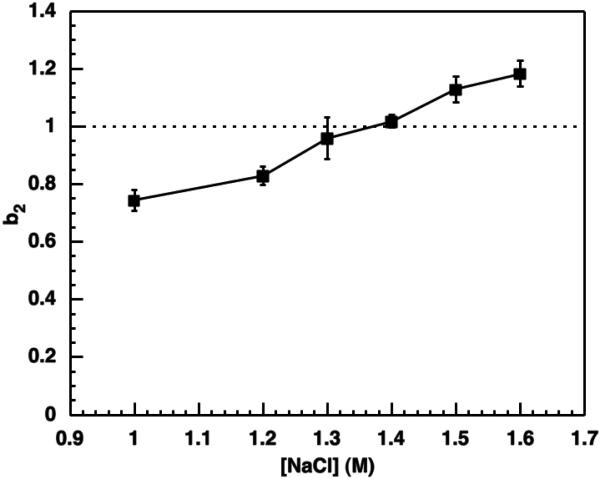
The virial coefficients of GI obtained by SIC in the presence of PEG near the polymorph transition are shown in Figure 3. Under these conditions the virial coefficient values are all positive, indicating that the net interaction is repulsive. The virial coefficients are normalized relative to the hard-sphere contribution (eq. 3), and the values obtained under these solution conditions indicate interactions that are on the order of the hard-sphere interactions, although the net effect of increasing NaCl concentration is to increase b2 consistently.
Figure 3.
Normalized osmotic second virial coefficients measured by SIC. All solutions contained 4% (w/v) PEG 10 kDa in 10 mM MES at pH 6.5. Error bars are based on triplicate measurements. The solid line is a guide to the eye, and the dotted line indicates the hard-sphere interactions.
Interestingly, the measured virial coefficients are all positive (repulsive) even though it is possible to produce crystals under these solution conditions. George and Wilson70 reported an empirical correlation of slightly negative (attractive) virial coefficients with an increased propensity for protein crystallization (“crystallization slot”). Observations of crystal growth outside the “crystallization slot”, such as that seen here, have been reported previously and appear to occur most often with larger proteins.39,71
Arakawa and Timasheff,72 using high-precision densimetric measurements, found that, for lysozyme and bovine serum albumin, there was an increase in the protein hydration at 1 M NaCl compared to that for a no-salt solution, by as much as 0.2-0.4 g water per g protein. Shulgin and Ruckenstein73 also calculated the preferential interaction parameter with salts using Kirkwood-Buff theory, showing that excess hydration of proteins with increasing salt concentration is theoretically consistent, although preferential hydration is cited as resulting in the salting-out of the protein. The increased hydration can be manifested as a steric layer of water on the protein surface that can interfere with short-range interactions through repulsive hydration effects.74 Since short-range interactions are likely the dominant interactions necessary to induce nucleation and growth of GI crystals, the presence of a layer of water would be expected to inhibit such association. The net repulsive virial coefficients and increasingly repulsive trend with increasing salt concentration observed can be interpreted as an increase in the excluded-volume interactions due to the excess hydration of GI with increasing NaCl. Also, the high concentration of Na+ ions in this system may lead to ion binding to the protein surface, since at pH 6.5 GI has a net charge of -54e. Na+ ions binding to the surface are also likely to add to the hydration layer as Na+ ions are highly hydrated.21 However, resolving the relative contributions of these possible mechanisms requires more detailed investigation, which was undertaken here by molecular modeling of different kinds.
Modeling of Crystal Contacts
A protein crystal usually represents the dense phase of minimum free energy, and since solubilities of different crystal forms can be identical under certain solution conditions (Figure 2), the free energies would also be expected to be identical. This allows protein crystal polymorphs to act as a sensitive probe of protein interaction energetics.
The two crystal polymorphs of GI investigated here grow within different space groups. The energetics of the unique crystal contacts were calculated using the identified crystal structures to determine if they correlate with the experimentally observed polymorph stability differences. Table 3 shows characteristics of the three unique types of contacts present in the two crystal forms of GI, including the number of interacting residues, bridging waters and the contact area for each contact. Each unique contact is identified by its representative symmetry transformation. The polyhedral crystals (PDB ID: 2GLK) are formed by a single unique contact (7546) that is repeated eight times for each GI tetramer. The rectangular crystals (PDB ID: 1OAD) exhibit two unique contacts, where one (4565) occurs four times and the other (2666) twice for each tetramer. Comparing the total number of residues involved in a crystal contact combined with the contact area suggests that the rectangular crystals would be expected to exhibit a stronger short-range interaction free energy per tetramer, thus suggesting the possibility of increased stability over polyhedral crystals at moderate to high salt.
Table 3.
Crystal contacts and associated interaction energies. Identified are the number of contact residues, salt bridges, bridging waters, the contact area as well as the calculated individual crystal contact short-range and electrostatic interaction free energies (at 1.3 M NaCl) and combined per tetramer interaction free energy, in units of kT.
| Crystal | Contact IDa | # residues | # salt-bridges | # waters | Contact area (Å2) | Interaction energies | ||
|---|---|---|---|---|---|---|---|---|
| short-range | electrostatic tetramerb | |||||||
| Polyhedral | 7546 (8) | 11 | 2 | 8 | 461 | −10.7 | 0.45 | −82.0 |
| Rectangle | 4565 (4) | 17 | 5 | 12 | 724 | −17.2 | 0.23 | −157.5 |
| Rectangle | 2666 (2) | 38 | 9 | 24 | 1885 | −45.1 | 0.29 | |
NMMM symmetry transformations are given where N is the serial number of the symmetry operator, and MMM is the concatenated cell translations along x,y,z with respect to the base 555. The numbers in parentheses indicate the coordination numbers of the contacts.
The combined tetramer interaction free energy accounts for all contacts made per tetratmer, i.e., eight for the 7546 contact, four for the 4565 and two for the 2666 contact.
Figure 4 shows the short-range interaction energy potentials, calculated using the hybrid molecular mechanics method, as a function of the gap distance in the absence of hydrating water, where the gap distance is defined as the protein center-to-center distance relative to the crystallographic contact position. The well minima (Table 3), all near a zero gap distance, exhibit strong attractive interaction energies for each contact. The ratio of the well minimum to crystal contact area is about 0.023 kT/Å2 for each of the contacts and consistent with the range of values reported previously.49
Figure 4.
Calculated short-range interaction free energies between two GI molecules at crystal contacts using only the non-hydrogen protein atoms. Gap distance represents the distance relative to the crystallographic position along a line connecting the protein centers. The polyhedral crystal (7456) interaction energy is shown in blue. The two rectangular crystal interaction energies are shown in green and red for the 4565 and 2666 crystal contacts, respectively.
To estimate the interaction free energy of the GI crystal packing molecule (tetramer) within each crystal form, additivity of the crystal contact interaction free energies due to short-range and electrostatic contributions was assumed. The results indicate that under conditions of high ionic strength, where short-range interactions are dominant, the rectangular crystals would be expected to be more stable, as is indeed observed experimentally. However, the calculated total tetramer interaction free energies (including the small electrostatic contributions) show an almost two-fold higher free energy change for the rectangular crystals than for the polyhedral crystals, without accounting for water, and the small electrostatic contribution does not change enough for this conclusion to change much as a function of NaCl concentration. In fact, the interaction free energies shown were calculated at the experimental point of transition in stability, 1.3 M NaCl, where the total tetramer interaction free energies of the crystal polymorphs are expected to be equal, indicating a pronounced discrepancy between the calculations and experimental observations.
The present short-range interaction model can potentially capture two opposing roles of water in crystal contacts. Interfacial water molecules can strengthen attraction by promoting better packing (enhanced complementarity) of the interface, or they can disrupt a contact interface by steric interference. Figure 5 reveals both possibilities for the 2666 crystal contact. Addition of crystallographic waters results in a significant strengthening of the crystal contact interaction free energy relative to that for the protein alone. The enhancement of the interaction free energy of the contact with crystallographic water is likely due to the contribution of the water molecules, which do not impact the complementarity, to short-range attraction. Although the polyhedral contact shows similar behavior to the 2666 contact, the 4565 contact does not, with the interaction free energy instead left unchanged by the presence of crystallographic water molecules.
Figure 5.
Calculated short-range interaction free energies between two GI molecules at a rectangular crystal contact (2666). Gap distance represents the distance relative to the crystallo-graphic position along a line connecting the protein centers. Solid red: interaction without water; solid blue: interaction including all crystallographic waters; dashed blue: interaction between fragments with bound water determined using MD for the contact pair; dashed red: interaction between fragments with bound water determined using MD for each isolated fragment.
Interaction free energies calculated for the various protein contacts accounting for protein hydration as in Figure 5 are summarized in Table 4. In the rectangular crystal contact 4565, inclusion of water based on the MD simulations (infinite separation or within the crystal contact) almost completely offsets the direct protein-protein interaction free energy (which was already >2.5 times less favorable than for the 2666 contact), giving the contact a calculated interaction free energy close to zero. In contrast, inclusion of the crystallographic water is calculated as stabilizing or enhancing the 2666 contact but has almost no effect on the 4565 contact. We may therefore infer that the 4565 contact provides only a small contribution to the assembly of rectangular crystals. Rectangular crystals likely nucleate as a result of the formation of adjacent 2666 contacts, hence the loss of symmetry, whereas growth along the 4565 contact may occur after the nucleation event, possibly by the formation of layers of tetramers associated by 2666 contacts. This may also explain the experimental observation of the rectangular crystals growing as flat plates in two dimensions prior to any substantial growth in the third dimension.
Table 4.
Calculated individual crystal contact short-range interaction free energies, in units of kT.
| Conditions | Polyhedrala | Rectangular (4565) | Rectangular (2666) |
|---|---|---|---|
| No water | −10.7 | −17.2 | −45.1 |
| Crystallographic water | −19.9 | −17.1 | −55.0 |
| MD independentb | −1.8 | −0.7 | −7.0 |
| MD in-contact | −9.7 | −0.4 | −34.5 |
Polyhedral crystals have only one unique contact, 7546.
Water is included for each protein fragment simulated independently.
Calculation of the interaction free energy between two fragments that included water molecules identified by MD simulations of independent fragments (infinite separation; Supporting Information Figure 1) provides an example of the ability of water molecules to block contact formation. This is clearly due to the presence of the water molecules because the two fragments are able to approach one another only to a gap distance of approximately a water molecule's diameter. Comparing the calculated short-range interaction free energies that include MD “bound” water (Table 4) suggests that dehydration of the interacting protein surfaces would be required prior to contact formation. However, including only the water within the first hydration layer from the simulations shows cases of both an increased strength of interaction and a destabilization of the short-ranged protein-protein interactions, when simulated with the fragments held in their contact positions, which depended on the contact being investigated.
Table 5 shows the calculated total interaction free energy of a tetramer interacting with all of its nearest neighbors in each crystal form. Again the influence of various levels of hydration can be seen. When crystallographic water is included, the interaction free energies are seen to increase significantly in magnitude for the polyhedral crystals, giving a difference in calculated contact energies between the two crystal forms of less than 12%. Finally, including the water based on the MD simulations (simulated in-contact) significantly reduces the magnitude of the interaction free energies compared to those for the crystallographic waters; however, the total interaction free energies are almost the same in magnitude (<10% difference), as is expected at the crossover point.
Table 5.
Calculated total (combined short-range and electrostatic) crystal interaction free energies, in units of kT.
| Conditions | Polyhedrala | Rectanglea |
|---|---|---|
| No water | −82 | −158 |
| Crystallographic water | −155 | −177 |
| MD waterb | −74 | −69 |
Polyhedral crystals include electrostatic energies of 0.45 kT for each of the eight identical contacts and rectangular crystals include 0.23 kT for each of the four 4565 contacts and 0.29 kT for each of the two 2666 contacts, all determined at 1.3 M NaCl.
MD water describes only the scenario where the water molecules were identified with the protein fragments held in their crystal contact positions throughout the simulations.
There are two cases for which the free energy and enthalpy of crystallization of GI have been measured experimentally,9,75 although in both cases these quantities were obtained for the polyhedral crystal form only. Vuolanto et al.9 measured the free energy of crystallization ΔGcryst = –15.6 kT and ΔHcryst = –48.4 kT in (NH4)2SO4 and MgSO4 solutions. These values were found to be insensitive to the cation type. Sleutel et al.75 obtained DHcryst = –63.0 kT and ΔGcryst = –9.4 kT in 2% (w/v) PEG 1000 with MgCl2. In 5% (w/v) PEG 1000 with MgCl2, the values found were ΔHcryst = –58.5 kT and DGcryst = –13.8 kT. The similar order-of-magnitude of ΔHcryst in very different systems, (NH4)2SO4 vs. PEG 1000 and MgCl2, suggests that about the same magnitude might be expected in the PEG 10 kDa, NaCl system used here.
Our calculations cannot be readily modified to separate the enthalpic and entropic components of the interaction free energy, but comparisons with experimental ΔGcryst are possible. The limitations of our method notwithstanding, the calculated interaction free energy in polyhedral crystal form ranges between about –10 and –20 kT, depending on how water is treated, while the experimental values range between about –10 and –16 kT;9,75 the similarities in the magnitude and range are encouraging. Although the lack of experimental data renders impossible even the above coarse level of comparison for the rectangular crystal form, we can infer plausible trends based on the size of the interfaces. Given the larger contact areas involved in the rectangular crystal form, it is plausible that the dehydration penalties are high, effectively increasing the energetic cost of crystallization. However, the same extensive contact also favors direct protein-protein interactions (cf. magnitudes of contacts in Table 4). Thus under low-salt conditions, dehydration could limit formation of the rectangular crystal form, effectively favoring the polyhedral form, while with increasing salt and hence also dehydration, the balance is tipped in favor of the rectangular form. We therefore consider studies with explicit salt in the simulations to probe these suggestions.
Water Affinity with NaCl
Although the role of water appears to be a major factor determining polymorph selection, the calculations thus far have not accounted for the real experimental system of a high NaCl concentration or the presence of PEG. Accounting for PEG in a MD simulation is computationally unfeasible; therefore, effects of different concentrations of only NaCl on water affinity were performed. The influence of PEG on crystallization is assumed to be due solely to depletion forces. Since the systems are all in the dilute PEG regime we can apply the Asakura-Oosawa76 model to estimate the contribution of PEG. Using the 4% PEG 10 kDa system, the contribution to the interaction free energies would be approximately −1 kT, and therefore is likely to have a minimal effect on the crystal contact free energies. Also, since the PEG concentration is constant, there should be no effect of protein surface hydration changes due to PEG.
To account better for the behavior of the experimental system, simulations to determine the water affinity (η) as a function of NaCl concentration were performed at 0, 1 and 2 M NaCl, with each relevant protein fragment simulated independently. To ensure that the simulations were long enough to allow for equilibration of the ions, the diffusion coefficients of Na+ and Cl– were calculated using the mean squared displacement over the full 4 ns production runs (Supporting Information Table 1) and compared to reported values,77–79 where all ions were were used for the calculations. There is appreciable variability among diffusion coefficients determined by simulation, although general trends can be compared. The simulated diffusion coefficient values found at 2 M are consistently smaller than at 1 M, in agreement with reported values.77–79 The diffusion coefficient of Cl– is observed to be consistently larger than that of Na+, also in agreement with reported simulation values.
The ion displacement data also allowed potentially bound ions to be identified from time periods where there was essentially no migration. Once identified, those ions were visually checked using VMD60 to determine the location of the ion relative to the protein fragment of interest. Interestingly, Na+ ions were often found to be bound to the protein surface for various periods of time (as long as ~ 0.5 ns) whereas Cl– ions were not found bound to the surface; rather, they were often found associated with Na+ ions that were bound to the surface. Friedman et al.77 observed similar times for Na+ near aspartate residues. The periods for which Cl– ions were found associated with Na+ ions were considerably shorter than those for which Na+ ions were associated with the protein (tens of picoseconds). Na+ ions were most often observed associated with the carboxylate oxygens present on the surface, regardless of the fragment.
The results for the different protein fragments at multiple NaCl concentrations show that binding of Na+ ions to GI can occur, and can modify the protein’s hydration layer locally. Experimental results shown earlier demonstrate that the polyhedral crystals are the more stable crystals at lower NaCl concentrations, regardless of the PEG concentration. The influence of NaCl concentration on the water affinity for the two representative fragments of the polyhedral crystals, as given by η (eq. 7), is shown in Table 6. The loss of water due to increasing NaCl concentration is expected, since ions can interfere with water association with the protein as well as affecting the overall ordering of water due to ion hydration. However, the gain of water molecules on the protein surface is contrary to this argument. The location of water molecules that could be identified as gained with an increase in NaCl concentration was therefore determined.
Table 6.
Number of water molecules predicted by MD simulations to be lost or gained on the protein surface as a function of NaCl concentration changes in polyhedral crystal fragmentsa.
| [NaCl] change [M] | Conserved | Lost | Gained | |
|---|---|---|---|---|
| Fragment 1 | 0→1 | 68 | 2 | 0 |
| 0→2 | 64 | 3 | 7 | |
| 1→2 | 62 | 4 | 9 | |
| Fragment 2 | 0→1 | 85 | 8 | 8 |
| 0→2 | 78 | 9 | 8 | |
| 1→2 | 84 | 7 | 5 | |
Gained water required η at lower NaCl concentration to be < 2 and Δη > 0.2.
As an example, Figure 6 shows the location of the seven gained waters in a polyhedral fragment for an increase in NaCl concentration from 0 to 2 M. Also included is a Na+ ion that was identified by the methods described previously as being bound to the protein surface. In this case the Na+ ion is found associated with the carboxylate group of an aspartate residue, as anticipated.21 The presence of the water surrounding the Na+ ion indicates that even though the ion is bound to the protein surface, it retains most of its own hydration shell. It is this hydration force that has been invoked to explain the cloud-point behavior of lysozyme as a function of salt type.80 The example shown here is not specific for this fragment, and in fact the simulations indicate that almost all of the water that is gained by including Na+ ions is associated with Na+ ions “bound” to carboxylate groups. In the polyhedral crystals there are only 11 residues involved in each crystal contact, and of those, three are aspartates and one is a glutamate, all of which could be expected to associate with Na+ ions and have the potential to inhibit contact formation. The rectangular crystal contact 4565 has 17 residues, of which two are aspartates and two are glutamates. The 2666 contact has 38 residues of which three are aspartates and 11 are glutamates. The ion binding of Na+ to the protein surface with a mostly intact hydration layer is likely to influence the contact formation for all three contacts, but the polyhedral crystal contacts are likely to be more strongly affected with the limited number of contact residues involved.
Figure 6.
Polyhedral crystal fragment 1 with bound water gained when the NaCl concentration was increased from 0 to 2 M. The side chain of aspartate 57 is shown with a bound Na+ ion (yellow) and gained water molecules (red).
Figure 7 shows the van der Waals surface of the tetramer of GI. The residues highlighted in red are the Asp and Glu residues at which water was predicted to be gained due to Na+ binding when the NaCl concentration was increased from 0 to 1 M. The residues in blue show the corresponding Asp and Glu residues for an NaCl concentration increase from 1 to 2 M. Finally, the Asp 80 residues that were observed to bind Na+ ions at 2 M NaCl to the polyhedral crystal fragment surface are shown in yellow. These residues were singled out from the other residues binding Na+ ions at 2 M NaCl because Asp 80 is a residue involved in the polyhedral crystal contact that forms two salt bridges to an adjacent tetramer. There are residues involved in the rectangular 4565 crystal contact that are also seen to bind Na+ ions, but the contribution to the overall tetramer interaction free energy of this contact is much smaller than that of the 2666 contact. Although there are a number of potential Na+ binding sites in the 2666 contact, none was found actually to bind Na+ ions, possibly due to the local protein environment. As mentioned earlier, when rectangular crystals are grown they are normally visible as flat plates growing in the z-direction, which is the same direction that the 2666 contact would promote. The rectangular crystals grow much more slowly in the third dimension.
Figure 7.
Van der Waals surface of a GI tetramer. Colored residues are Asp, Glu and the C-terminus on each monomer, determined by MD simulations to bind Na+ ions and add water upon addition of 1 M (red) or 2 M (blue) NaCl. Asp 80, colored yellow, is where Na+ ions bind and add water at 2 M NaCl and is identified as a residue involved in salt-bridging within the polyhedral crystal contact.
In the case of GI the modeling suggests that an increase in the NaCl concentration far beyond the range for normal electrostatic screening contributes to the modification of the crystal polymorph stability. Comparison of common monovalent cations suggests that only Li+ may have a higher affinity than Na+ for the protein surface,81 since it is a smaller, more strongly hydrated cation than sodium, but the effects of lithium salts have been described as unusual82 and the hydration energy of Li+ is sufficiently high to make removing water from its first hydration layer difficult. Conversely, K+ and larger monovalent cations would have reduced affinity for the protein surface and requiring higher concentrations to affect crystal stability. The preference for Na+ over K+ at the protein surface has been observed for multiple proteins.21 Changing the cation from monovalent to divalent would be expected to increase the stability of the polyhedral crystals due to the ion’s molecular size and hydration free energy, which was observed for GI when the salt was changed from (NH4)2SO4 to MgSO4,9 where rectangular crystals were never observed.
Conclusions
This work provides both experimental determination and modeling insights for the physico-chemical properties that control GI crystal polymorph selection. Crystal polymorph selection was experimentally found to be influenced by high NaCl concentrations in the presence of PEG, as was shown by two different methods, where a stable polyhedral crystal was observed at low ionic strengths and a rectangular crystal was observed at very high ionic strengths, specifically > 1.3 M. Selection is likely due to ion binding and changes in protein surface hydration, with repulsive osmotic second virial coefficients supporting this theory. The observation of crystallization under net repulsive conditions stands in contrast to the emipirial correlation of slightly negative (attractive) virial coefficients described by George and Wilson.70
Through the application of multi-scale models a controlling mechanism of polymorphic crystal selection was identified as a combination of cation concentration-dependent surface ion binding coupled with local hydration. Through the use of the hybrid LH/LJ method the calculated short-ranged interaction free energies were observed to be quite sensitive to the approach used to approximate protein surface hydration. When concentrating on very specific protein-protein orientations, such as a crystal contact, accurate accounting for the effects of hydration and salt effects was deemed critical, with the quasi-chemical approach offering a satisfactory method to account for protein surface hydration.
While preferential hydration of the protein surface with increasing salt concentration is presented as an explanation of salting-out behavior,73,83 the presence of water at the protein surface can also sterically inhibit protein self-association. Although increasing salt is more often associated with the screening of electrostatic interactions, the results presented here identify an additional role, that of ion binding combined with the presence of a largely intact ion hydration layer, which may help to account for the salting-in behavior of some proteins with increasing salt. Additionally, ion binding to the protein surface can result in other changes in protein-protein interactions, specifically formation of salt bridges and inhibition of contact formation. The experimental methods and observations described here provide a practical approach for the identification of solution conditions relevant for polymorph selection for applications such as refinement of crystallization conditions and protein formulation, while the modeling approach offers a method of quantitative identification of a mechanism of crystal polymorph selection.
Supplementary Material
Acknowledgements
We are grateful to Uttamkumar Samanta and Brian Bahnson for their assistance in obtaining x-ray diffraction data for our crystals. We also acknowledge M. Hamsa Sundaram and Mike Paulaitis for useful discussions and suggestions during the modeling part of this work. C.M.G. gratefully acknowledges support through a NIH Chemistry-Biology Interface Training Grant and through an Alfred P. Sloan Minority Fellowship. The research was supported by the National Science Foundation under grant number BES-0519191.
Footnotes
Supporting Information Available
Exemplary cartoon representation of protein fragments as used in the MD simulations and a table comparing the calculated and reported ion diffusion coefficients determined from the simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hekmat D, Hebel D, Weuster-Botz D. Chem. Eng. & Tech. 2008;31:911–916. [Google Scholar]
- 2.Pechenov S, Shenoy B, Yang MX, Basu SK, Margolin AL. J. Control. Rel. 2004;96:149–158. doi: 10.1016/j.jconrel.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Basu SK, Govardhan CP, Jung CW, Margolin AL. Expert Opn. Biol. Therapy. 2004;4:301–317. doi: 10.1517/14712598.4.3.301. [DOI] [PubMed] [Google Scholar]
- 4.Shire SJ, Shahrokh Z, Liu JJ. Pharm. Sci. 2004;93:1390–1402. doi: 10.1002/jps.20079. [DOI] [PubMed] [Google Scholar]
- 5.Lafont S, Veesler S, Astier JP, Boistelle RJ. Cryst. Growth. 1994;143:249–255. doi: 10.1107/S0907444993014416. [DOI] [PubMed] [Google Scholar]
- 6.Lafont S, Veesler S, Astier JP, Boistelle RJ. Cryst. Growth. 1997;173:132–140. [Google Scholar]
- 7.Veesler S, Ferte N, Costes MS, Czjzek M, Astier JP. Cryst. Growth & Des. 2004;4:1137–1141. [Google Scholar]
- 8.Boistelle R, Astier JP, Marchismouren G, Desseaux V, Haser RJ. Cryst. Growth. 1992;123:109–120. [Google Scholar]
- 9.Vuolanto A, Uotila S, Leisola M, Visuri KJ. Cryst. Growth. 2003;257:403–411. [Google Scholar]
- 10.Vivares D, Veesler S, Astier JP, Bonnete F. Cryst. Growth & Des. 2006;6:287–292. [Google Scholar]
- 11.Wlodawer A, Borkakoti N, Moss DS, Howlin B. Acta Cryst. B. 1986;42:379–387. [Google Scholar]
- 12.Kowacz M, Mukhopadhyay A, Carvalho AL, Esperanca JMSS, Romao MJ, Rebelo LP. N. CrystEngComm. 2012;14:4912–4921. [Google Scholar]
- 13.Crosio M-P, Janin J, Jullien MJ. Mol. Biol. 1992;228:243–251. doi: 10.1016/0022-2836(92)90503-c. [DOI] [PubMed] [Google Scholar]
- 14.Michalska K, Borek D, Hernandez-Santoyo A, Jaskolski M. Acta Cryst. D. 2008;64:309–320. doi: 10.1107/S0907444907068072. [DOI] [PubMed] [Google Scholar]
- 15.Simone S, Curcio E, Di Profio G, Ferraroni M, Drioli EJ. Membrane Sci. 2006;283:123–132. [Google Scholar]
- 16.Giffard M, Colloc'h N, Ferte N, Castro B, Bonnete F. Cryst. Growth & Des. 2008;8:4220–4226. [Google Scholar]
- 17.Tomčová I, Smatanová IK. J Cryst. Growth. 2007;306:383–389. [Google Scholar]
- 18.Asherie N, Jakoncic J, Ginsberg C, Greenbaum A, Stojanoff V, Hrnjez BJ, Blass S, Berger J. Crystal Growth Design. 2009;9:4189–4198. [Google Scholar]
- 19.Trakhanov S, Quiocho FA. Protein Sci. 1995;4:1914–1919. doi: 10.1002/pro.5560040925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trakhanov S, Kreimer DI, Parkin S, Ames GFL, Rupp B. Protein Sci. 1998;7:600–604. doi: 10.1002/pro.5560070308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vrbka L, Vondrasek J, Jagoda-Cwiklik B, Vacha R, Jungwirth P. Proc. Natl. Acad. Sci. USA. 2006;103:15440–15444. doi: 10.1073/pnas.0606959103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lund M, Vacha R, Jungwirth P. Langmuir. 2008;24:3387–3391. doi: 10.1021/la7034104. [DOI] [PubMed] [Google Scholar]
- 23.Pashley RM. Adv. Colloid Interface Sci. 1982;16:57–62. [Google Scholar]
- 24.Pashley RM. J. Colloid Interface Sci. 1981;83:531–546. [Google Scholar]
- 25.Pashley RJ. Colloid Interface Sci. 1981;80:153–162. [Google Scholar]
- 26.Pashley RM, Israelachvili JN. J. Colloid Interface Sci. 1984;97:446–455. [Google Scholar]
- 27.Bhosale SH, Rao MB, Deshpande VV. Microbiol. Rev. 1996;60:280–300. doi: 10.1128/mr.60.2.280-300.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chayen N, Akins J, Campbell-Smith S, Blow DM. J. Crystal Growth. 1988;90:112–116. [Google Scholar]
- 29.Chayen NE, Lloyd LF, Collyer CA, Blow DM. J. Crystal Growth. 1989;97:367–374. [Google Scholar]
- 30.Vivares D, Kaler EW, Lenhoff AM. Acta Cryst. D. 2005;61:819–825. doi: 10.1107/S090744490402949X. [DOI] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl. Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, Minor W. Macro. Crystallogr. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Berger BW, Blamey CJ, Naik UP, Bahnson BJ, Lenhoff AM. Cryst. Growth & Des. 2005;5:1499–1507. [Google Scholar]
- 34.Dumetz AC, Snellinger-O’Brien AM, Kaler EW, Lenhoff AM. Protein Sci. 2007;16:1867–1877. doi: 10.1110/ps.072957907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tessier PM, Johnson HR, Pazhianur R, Berger BW, Prentice JL, Bahnson BJ, Sandler SI, Lenhoff AM. Proteins. 2003;50:303–311. doi: 10.1002/prot.10249. [DOI] [PubMed] [Google Scholar]
- 36.Tessier PM, Lenhoff AM, Sandler SI. Biophys. J. 2002;82:1620–1631. doi: 10.1016/S0006-3495(02)75513-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePhillips P, Lenhoff AM. J. Chrom. A. 2000;883:39–54. doi: 10.1016/s0021-9673(00)00420-9. [DOI] [PubMed] [Google Scholar]
- 38.Kozak MJ. Appl. Crystallogr. 2005;38:555–558. [Google Scholar]
- 39.Bonnete F, Vivares D. Acta Cryst. D. 2002;58:1571–1575. doi: 10.1107/s090744490201418x. [DOI] [PubMed] [Google Scholar]
- 40.Moreels E, Deceuninck W, Finsy RJ. Chem. Phys. 1987;86:618–623. [Google Scholar]
- 41.Paliwal A, Asthagiri D, Abras D, Lenhoff AM, Paulaitis ME. Biophys. J. 2005;89:1564–1573. doi: 10.1529/biophysj.105.065284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimm BH. J. Chem. Phys. 1948;16:1093–1099. [Google Scholar]
- 43.Velev OD, Kaler EW, Lenhoff AM. Biophys. J. 1998;75:2682–2697. doi: 10.1016/S0006-3495(98)77713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katz AK, Li XM, Carrell HL, Hanson BL, Langan P, Coates L, Schoen-born BP, Glusker JP, Bunick GJ. Proc. Natl. Acad. Sci. USA. 2006;103:8342–8347. doi: 10.1073/pnas.0602598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramagopal UA, Dauter M, Dauter Z. Acta Cryst. D. 2003;59:868–875. doi: 10.1107/s0907444903005663. [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick KJ. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 47.Connolly ML. J. Molec. Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 48.Neal BL, Lenhoff AM. AIChE J. 1995;41:1010–1014. [Google Scholar]
- 49.Asthagiri D, Neal BL, Lenhoff AM. Biophys. Chem. 1999;78:219–231. doi: 10.1016/s0301-4622(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 50.Jorgensen WL, Tiradorives JJ. Amer. Chem. Society. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 51.Roth CM, Neal BL, Lenhoff AM. Biophys. J. 1996;70:977–987. doi: 10.1016/S0006-3495(96)79641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asthagiri D, Paliwal A, Abras D, Lenhoff AM, Paulaitis ME. Biophys. J. 2005;88:3300–3309. doi: 10.1529/biophysj.104.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zubay G. Biochemistry. Addison-Wesley; New York: 1984. [Google Scholar]
- 54.Stryer L. Biochemistry. W.H. Freeman and Co.; New York: 1988. [Google Scholar]
- 55.Yoon BJ, Lenhoff AM. J. Comp. Chem. 1990;11:1080–1086. [Google Scholar]
- 56.Paulaitis ME, Pratt LR. Adv. Protein Chem. 2002;62:283–310. doi: 10.1016/s0065-3233(02)62011-x. [DOI] [PubMed] [Google Scholar]
- 57.Makarov VA, Andrews BK, Pettitt BM. Biopolymers. 1998;45:469–478. doi: 10.1002/(SICI)1097-0282(199806)45:7<469::AID-BIP1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 58.Lin B, Wong K-Y, Hu C, Kokubo H, Pettitt BM. J. Phys. Chem. Lett. 2011;2:1626–1632. doi: 10.1021/jz200609v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten KJ. Comp. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten KJ. Molec. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.MacKerell AD, Banavali N, Foloppe N. Biopolymers. 2000;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 62.Neria E, Fischer S, Karplus MJ. Chem. Phys. 1996;105:1902–1921. [Google Scholar]
- 63.Miyamoto S, Kollman PA. J. Comp. Chem. 1992;13:952–962. [Google Scholar]
- 64.Darden T, York D, Pedersen LJ. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 65.Hoover WG. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 66.Hamiaux C, Perez J, Prange T, Veesler S, Ries-Kautt M, Vachette PJ. Mol. Biol. 2000;297:697–712. doi: 10.1006/jmbi.2000.3584. [DOI] [PubMed] [Google Scholar]
- 67.Asherie N. Methods. 2004;34:266–272. doi: 10.1016/j.ymeth.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 68.Chernov AA. Phys. Rep. 1997;288:61–75. [Google Scholar]
- 69.Bloustine J, Virmani T, Thurston GM, Fraden S. Phys. Rev. Lett. 2006;96:087803. doi: 10.1103/PhysRevLett.96.087803. [DOI] [PubMed] [Google Scholar]
- 70.George A, Wilson WW. Acta Cryst. D. 1994;50:361–365. doi: 10.1107/S0907444994001216. [DOI] [PubMed] [Google Scholar]
- 71.Blouwolff J, Fraden SJ. Cryst. Growth. 2007;303:546–553. [Google Scholar]
- 72.Arakawa T, Timasheff SN. Biochemistry. 1982;21:6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 73.Shulgin IL, Ruckenstein E. Biophys. Chem. 2005;118:128–134. doi: 10.1016/j.bpc.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Israelachvili JN. Intermolecular & Surface Forces. Second ed. Academic Press Inc.; London: 2006. [Google Scholar]
- 75.Sleutel M, Willaert R, Gillespie CM, Evrard C, Wyns L, Maes D. Cryst. Growth & Des. 2009;9:497–504. [Google Scholar]
- 76.Asakura S, Oosawa FJ. Poly. Sci. 1958;33:183–192. [Google Scholar]
- 77.Friedman R, Nachliel E, Gutman M. Biophys. J. 2005;89:768–781. doi: 10.1529/biophysj.105.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Uchida H, Matsuoka M. Fluid Phase Equil. 2004;219:49–54. [Google Scholar]
- 79.Weerasinghe S, Smith PE. J. Chem. Phys. 2003;119:11342–11349. [Google Scholar]
- 80.Park EJ, Bae YC. Biophys. Chem. 2004;109:169–188. doi: 10.1016/j.bpc.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 81.Hess B, van der Vegt NF. A. Proc. Natl. Acad. Sci. USA. 2009;106:13296–13300. doi: 10.1073/pnas.0902904106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomas AS, Elcock AH. J. Amer. Chem. Society. 2007;129:14887–14898. doi: 10.1021/ja073097z. [DOI] [PubMed] [Google Scholar]
- 83.Arakawa T, Timasheff SN. Biochemistry. 1984;23:5912–5923. doi: 10.1021/bi00320a004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.