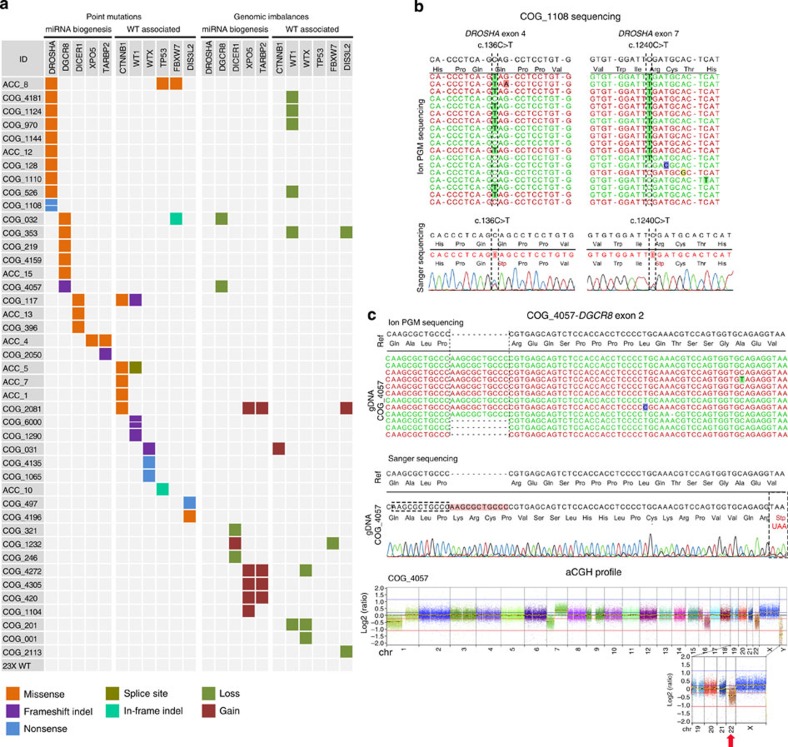
Figure 2. Mutation spectrum of WTs.
(a) Mutations of the miRNA core processing genes (miRNA biogenesis) and WT-associated genes (WT associated) in 66 fresh-frozen WT samples. Only the genes that were affected by point mutations in at least one sample are shown. Point mutations and indels (left panels) were identified by targeted parallel sequencing, and genomic imbalances (right panels) were detected by aCGH. aCGH data were obtained for 53 samples (2 from ACC and 51 from COG) from a previous study of the group. The coloured squares refer to the corresponding type of point mutation (missense, splice site, frameshift indel, in-frame indel and nonsense) or genomic imbalance (loss and gain). A detailed description of each mutation is provided in Table 1; Supplementary Tables 4 and 5. (b) DROSHA nonsense mutations (c.136C>T; p.Q46* and c.1240C>T; p.R414*) identified in a single patient (COG_1108) by targeted parallel sequencing (upper panels) and validated by capillary Sanger sequencing (bottom panels). (c) Mutations identified in the DGCR8 gene. The first panel depicts the 11-nt frameshift duplication identified by targeted parallel sequencing. The middle panel presents the validation by capillary sequencing and the translation of the mutated allele, highlighting the formation of a premature stop codon 62 nt downstream of the alteration. This patient (COG_4057) also presented a heterozygous loss of the entire chromosome 22 (aCGH profile—bottom panel), leading to the deletion of the wild-type DGCR8 allele. Owing to tumour heterogeneity and/or normal cell contamination, this aneuploidy is present in mosaic, resulting in ~30% of reads from Ion Torrent sequencing displaying the wild-type allele and in a log2 ratio value of −0.4 in aCGH analysis.