Table 2. Hydrogen-bond geometry (, ) for the crystal structures.
H-atom positions are normalized to average neutron-derived distances: CH = 1.089, NH = 1.015, OH = 0.993 (Allen Bruno, 2010 ▶).
| HA | D A | DHA | Symmetry code | |
|---|---|---|---|---|
| ACMINA (1:1) | ||||
| N2H2AO3 | 1.97 | 2.977(3) | 179 |
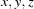
|
| N2H2BO7 | 1.92 | 2.889(3) | 161 |
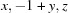
|
| O6H6AN3 | 1.65 | 2.636(3) | 177 |

|
| C2H2O6 | 2.29 | 3.369(3) | 174 |
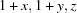
|
| C12H12O1 | 2.32 | 2.892(2) | 111 | Intramolecular |
| C17H17AO3 | 2.46 | 3.483(3) | 156 |
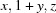
|
| C20H20BO1 | 2.42 | 3.471(3) | 164 |
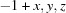
|
| ACMPAM (1:1) | ||||
| N2H2AO5 | 1.91 | 2.866(4) | 156 |
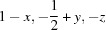
|
| N2H2BN3 | 2.32 | 2.722(4) | 102 | Intramolecular |
| O4H4O7 | 1.55 | 2.518(4) | 167 |
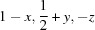
|
| C2H2O4 | 2.27 | 3.333(4) | 166 |
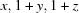
|
| C20H20AO1 | 2.49 | 3.507(4) | 156 |
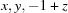
|
| C26H26O2 | 2.42 | 3.455(4) | 159 |

|
| ACMCPR (1:1) | ||||
| N2H2AO7 | 1.97 | 2.955(4) | 165 |
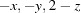
|
| O4H4O7 | 1.90 | 2.665(4) | 133 |
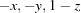
|
| C18H18BO6 | 2.18 | 3.094(5) | 140 |
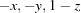
|
| C23H23ACl1 | 2.56 | 3.401(5) | 134 |
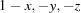
|
| C27H27BO5 | 2.08 | 2.972(4) | 138 |
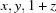
|
| ACMPABA (1:1) | ||||
| N2H2AO1 | 2.02 | 2.965(4) | 156 |
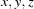
|
| N2H2BN2 | 2.24 | 3.215(5) | 163 |

|
| O7H7AO5 | 1.66 | 2.632(4) | 168 |
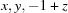
|
| C12H12O1 | 2.35 | 2.888(6) | 109 | Intramolecular |
| C15H15O2 | 2.40 | 3.458(6) | 166 |
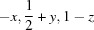
|
| C17H17AO3 | 2.42 | 3.493(6) | 172 |
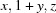
|
| C18H18AO3 | 2.38 | 3.437(6) | 164 |
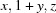
|
| C25H25O1 | 2.33 | 3.267(6) | 144 |

|
| ACMPPZ (1:0.5) | ||||
| N2H2AO4 | 2.51 | 3.226(4) | 128 |
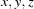
|
| N2H2AO5 | 1.63 | 2.619(3) | 166 |
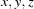
|
| N2H2BO4 | 1.80 | 2.798(3) | 172 |
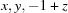
|
| C2H2O1 | 2.35 | 3.202(4) | 134 |
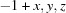
|
| C13H13O1 | 2.13 | 2.739(4) | 113 | Intramolecular |
| C17H17BO3 | 2.20 | 3.098(4) | 139 |
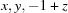
|
| C20H20AO4 | 2.34 | 3.055(4) | 122 |
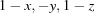
|
| C22H22AO5 | 2.33 | 3.362(4) | 158 |
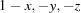
|
| C22H22BO2 | 2.34 | 3.020(4) | 119 |
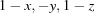
|
