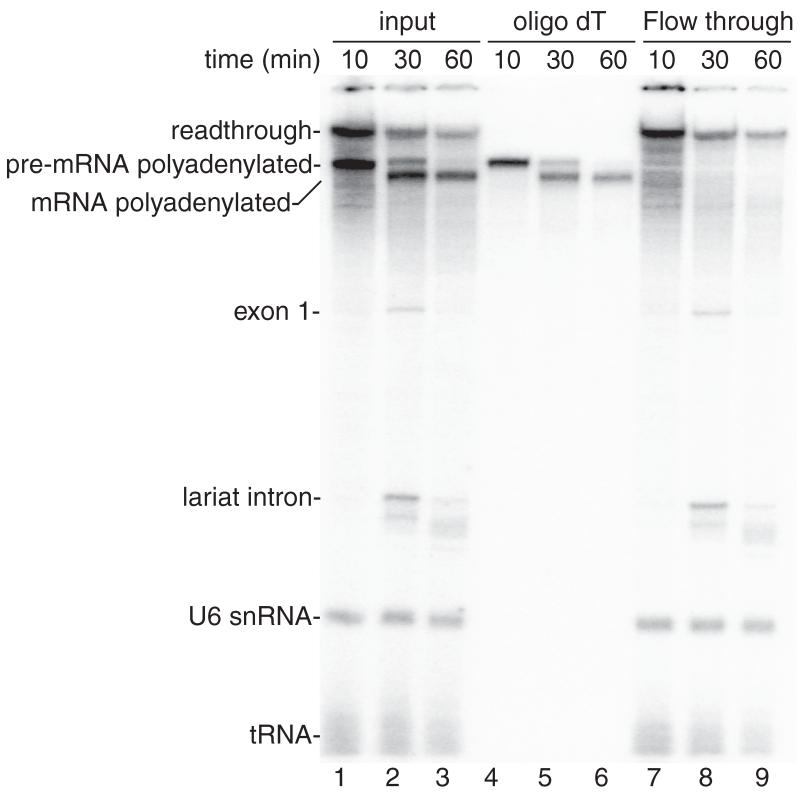
Figure 2.
Coupled RNAP II transcription/splicing/polyadenylation in vitro. (A) Structure of the DoF CMV-DNA template showing the pA signal. The sizes of the exons and intron are indicated. B. 32P-UTP and the CMV-DoF/BGH DNA template were incubated for 20 minutes to assemble a pre-initiation complex followed by addition of 32P-UTP, ATP, and CrPhos and continued incubation for 10 minutes. The readthrough transcript and polyadenylated pre-mRNA are generated by this time point (lane 1). Spliced polyadenylated mRNA is generated at the subsequent time points (lanes 2, 3). Total RNA from aliquots of the reactions shown in lanes 1-3 were isolated and passed through an oligo dT column. The polyadenylated pre-mRNA and mRNA bound to the oligo dT (lanes 4-6) whereas the readthrough transcript, exon 1, the lariat intron, U6 snRNA and tRNA are detected in the flow through from the oligo dT column (lanes 7-9).