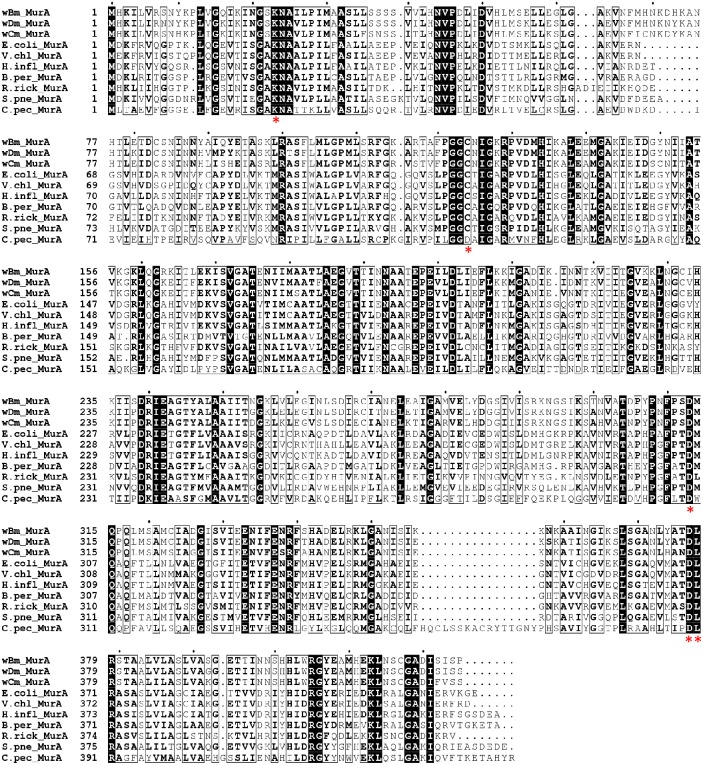
Figure 2. ClustalW analysis of the amino acid sequences of MurAs.
Sequences from various organisms were aligned (NCBI reference numbers are given in parenthesis). Wolbachia endosymbiont of B. malayi - wBm_MurA (YP_198570.1), Drosophila melanogaster - wDm_MurA (NP_966909), and Culex molestus - wCm_MurA (CDH88571); E. coli_MurA (WP_023568349), Vibrio cholerae - V.chl_MurA (WP_001887759), Haemophilius influenzae - H.infl_MurA (YP_005827986), Bordetella pertusis - B.per_MurA (WP_014906110), Rickettsia rickettsii - R.rick_MurA (WP_012151046), Streptococcus pneumoniae - S.pne_MurA (WP_023396463) and Chlamydia pecurum - C.pec_MurA (YP_008583337). Conserved motifs are marked and boxed, white letters with black background indicate identical amino acid positions/sequence from various organisms, red star shows the residues involved in the ligand interactions during catalysis. The alignment figure was generated using the ESPript 3.0 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).