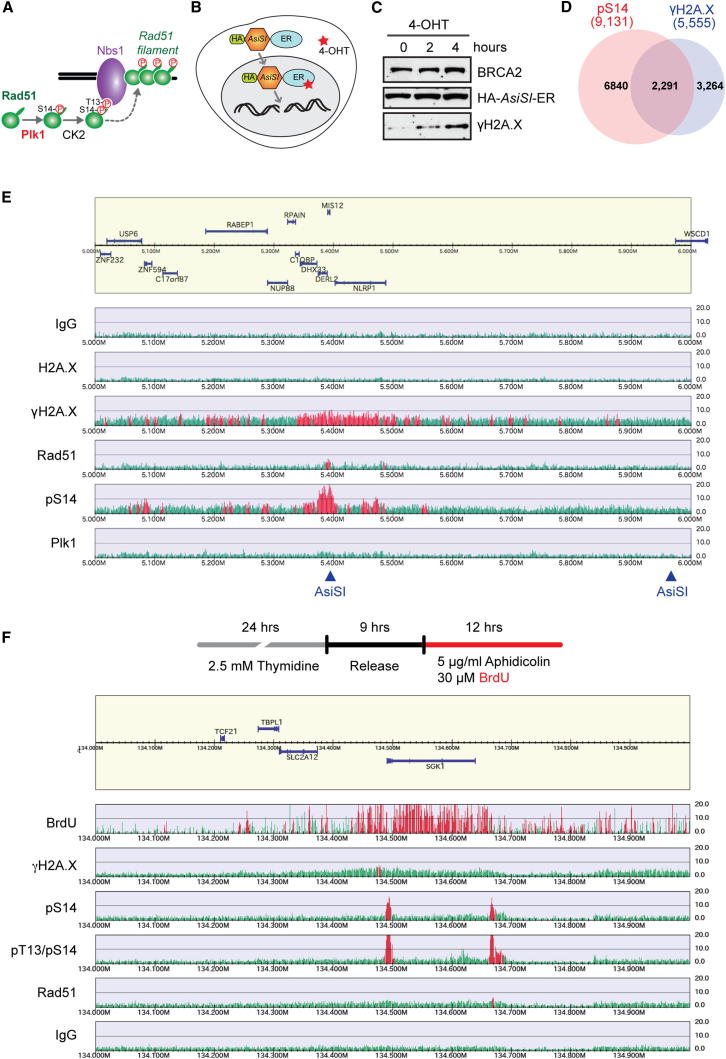
Figure 4.
Phosphorylated Rad51 Accumulates at Sites of DSB and Replicative DNA
(A) Model showing sequential Rad51 phosphorylation mediated by Plk1 and CK2, and recruitment of Rad51 to stressed DNA via phospho-dependent binding to Nbs1.
(B) Depiction of DSBs induction at AsiSI sites across the genome using U2OS cells stably expressing HA-AsiSI-ER fusion (U2OS AsiSI-ER).
(C) U2OS AsiSI-ER cells were treated with 0.3 mM 4-OHT for the indicated times, and HA-AsiSI-ER and γH2A.X in WCEs were detected using HA and γH2A.X antibodies, respectively.
(D) Venn diagram representing the number of colocalizing peaks for pS14 and γH2A.X quantified from genome-wide ChIP-seq results, using U2OS AsiSI-ER cells treated as in (C).
(E) Representative ChIP-seq profile of DNA-damage-responsive factors at a DSB site within chromosome 17, 5–6 Mb, using U2OS AsiSI-ER cells treated with 4-OHT for 4 hr. Arrowheads indicate locations containing the AsiSI target sequence (GCGATCGC).
(F) Representative ChIP-seq profile following replication stress within chromosome 6, 134–135 Mb, using HeLa cells treated with 5 μg/ml aphidicolin. Nascent DNA is labeled with the incorporation of BrdU.
See also Figure S4.