Figure 3.
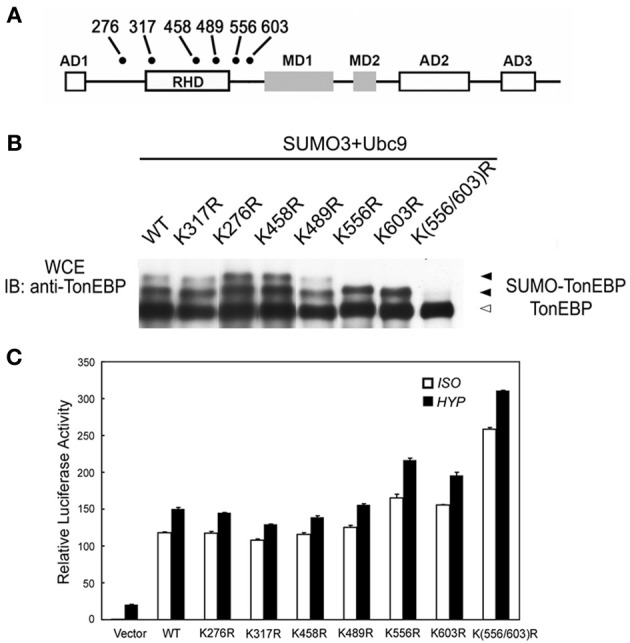
TonEBP is sumoylated on lysines 556 and 603. (A) Positions of lysine residues that fit the consensus sequence for sumoylation (ΨKxE: Ψ is a large hydrophobic residue; x is any residue) on a schematic primary structure of TonEBP. Rel homology domain (RHD), three activation domains (AD), and two modulation domains (MD) are shown. (B) HEK293 cells were transfected with wild type TonEBP (WT) or each of the TonEBP mutants shown along with SUMO3 and Ubc9. The cells were cultured for 4 h in hypertonic medium before immunoblot detection of TonEBP. Positions of TonEBP and sumoylated TonEBP (SUMO-TonEBP) are indicated. (C) HEK293 cells were co-transfected with a TonE-driven luciferase reporter plus empty expression plasmid (Vector) or expression plasmid containing WT or each of the TonEBP mutants as indicated. The cells were cultured for 4 h in isotonic (ISO) or hypertonic (HYP) medium before analysis of luciferase. Expression of luciferase activity is shown relative to isotonic vector control. K556R or K603R is significantly different from WT or K(556/603) both in ISO and HYP: p < 0.01. Mean + s.e.m., n = 6.
