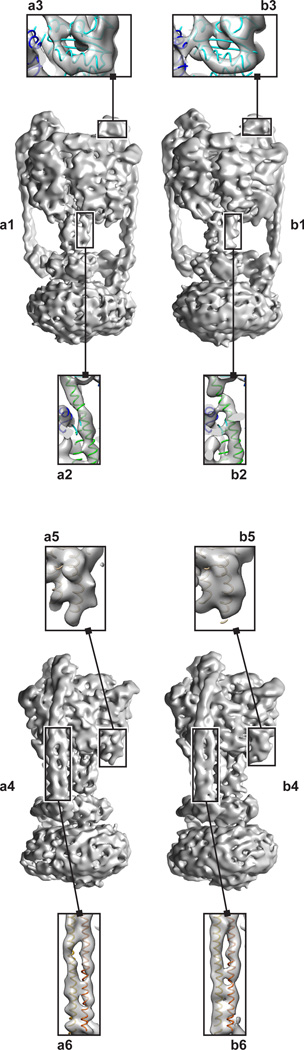
Figure 2.
3D map of the V/A-ATPase from T. thermophilus. A map reconstructed from a) automatically and b) manually picked particle images. The left panel (a1,b1) shows the V/A-ATPase from the front. Within this view, specific features highlight the resolvability of an α-helix from the central core, residues 1–37 of the D subunit, (a2,b2) versus a peripheral α-helix, residues 195–214 of the A subunit, (a3,b3). The right panel (a4,b4) shows the V/A-ATPase from the side. Within this view, specific features that highlight resolvability of a helix-turn-helix motif, residues 538–576 of chain A, (a5,b5) as compared to an α-helix in the stalk, residues 3–57 of the B subunit and residues 25–79 of the E subunit, (a6,b6).