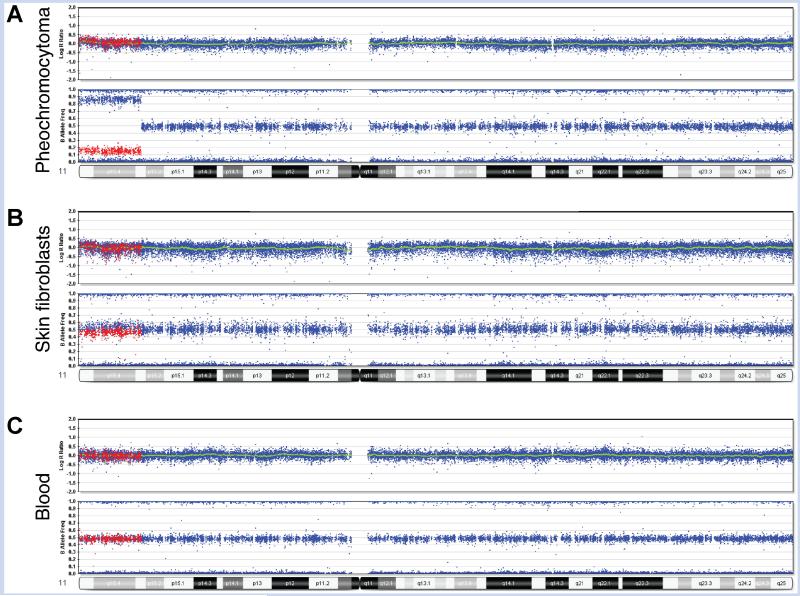
FIG. 2.
SNP array analysis data from multiple tissues of Patient 1. Illumina Human Quad 610 BeadChip results for chromosome 11. A: Pheochromocytoma demonstrates 70% mosaic isodisomy. B: Skin fibroblasts from hyperplastic leg demonstrate 5% mosaicism. C: Blood lymphocytes show no mosaicism. For illustration purposes, SNPs mosaic for heterozygosity and homozygosity of the minor allele in the pheochromocytoma are colored in red to demonstrate comparison of these same SNPs between the different tissues. In cells with normal levels of heterozygosity (peripheral blood), these red SNPs appear at the B allele frequency of 50%. In the cells with mosaic homozygosity, these red SNPs show a shift toward 0%, or an AA genotype (fibroblasts and tumor). Using this shift, the percent of cells with homozygosity was calculated as 5% in fibroblasts and 70% in tumor. In the tumor, heterozygous deletions in 8p12pter, 21q21.1qter, and 22q11.23qter were detected at lower percentages than the loss of heterozygosity (LOH) of 11p15.3pter, suggesting that the LOH was the primary event (data not shown).