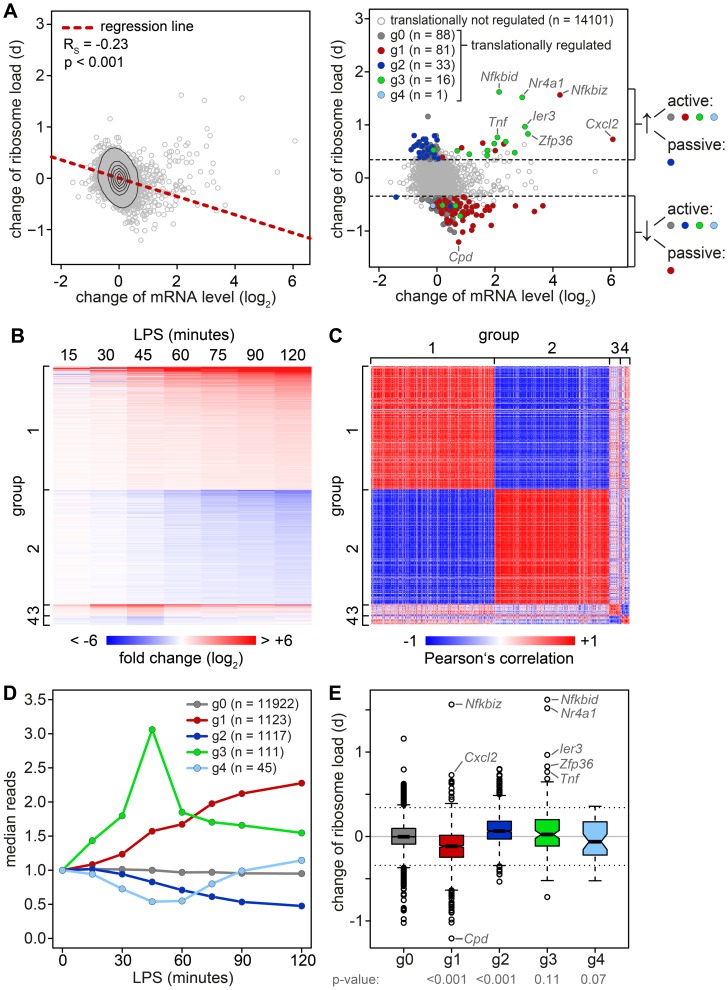
Figure 4. Relation between changes in mRNA levels and changes in translation.
(A) Left panel: Change of translation d (orthogonal distance from the regression line in Figure 3B) versus change of mRNA levels in RAW264.7 macrophages after 1 h of LPS treatment as quantified by microarray (n = 3). Right panel: Change of translation d versus change of mRNA levels, color-coded according to groups determined by RNASeq. (B) Groups of mRNAs with different response patterns during activation of RAW264.7 macrophages, as determined by RNASeq (n = 1). g1 and g2 contain mRNAs with a first significant maximum or minimum, respectively, at or after 1 h of LPS treatment (p<0.05 and log2(fold change) >0.5 or <0.5). g3 and g4 contain mRNAs with one significant maximum or minimum, respectively, before 1 h. Fold changes (log2) to the control are represented by the intensity of blue (negative) or red (positive). (C) Pair-wise Pearson's correlation coefficient of all mRNAs in g1–4. The strength of correlation is represented by the intensity of blue (negative) or red (positive). (D) mRNA expression patterns of the groups g0–4 as median counts normalized to the median of the control condition. g0 contains all mRNAs that do not show a significant change. (E) Box plot showing the change of translation d for each of the groups; p-values were determined by two-sided Wilcoxon rank sum test.