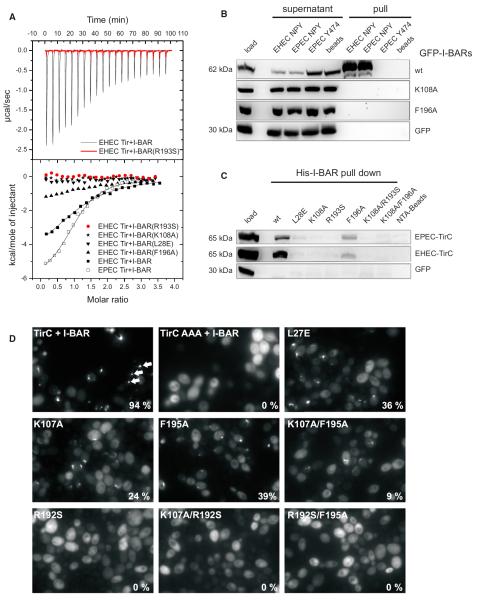
Figure 5. Interaction of Tir with Wild-Type and Mutant I-BAR Domains.
(A) ITC of the binding reaction of I-BARIRSp53 and four I-BAR point mutants with peptides derived from EHEC and EPEC Tir, comprising the NPY motif. The
upper panel shows considerable heat release upon titration of the wild-type I-BARIRSp53 (black) with the EHEC Tir-derived peptide, in contrast to the R193S mutant (red). The heat data, corrected for dilution, are plotted against the molar ratio of peptide molecules and I-BAR protomers in the lower panel. Sigmoidal curves typical of exothermic-binding reactions were obtained for the wild-type I-BARIRSp53 domain and the mutant F196A, but not for the R193S, K108A, and L28E mutants.
(B) Pull-down assays of Tir-derived peptides from lysates of mammalian cells ectopically expressing GFP-tagged I-BAR variants of human IRSp53 as indicated on the right. Shown are cell lysates before (load) and after (supernatant) pull-downs and precipitates (pull) of three different peptides as indicated above. EHEC Tir-derived peptides that bind to Nck instead of IRSp53 (Y474) as well as streptavidin beads were employed as bait-control, and GFP alone in the cell lysate was the prey-control.
(C) Pull-down assays of recombinant His-tagged I-BAR domains from lysates of COS7 cells ectopically expressing the cytoplasmic C termini of EPEC or EHEC Tir as indicated on the right. Shown is the cell lysate before the pull-down (load) and the precipitates (His-I-BAR pull-down) of wild-type I-BAR (wt) and six variants as indicated above. Note the weak binding to mutant F196A in accordance with the ITC measurements.
(D) Interaction of Tir with wild-type and mutant I-BAR domain of IRTKS in vivo. Yeast cells coexpressing μNS-TirC and wild-type or mutant GFP-tagged I-BAR domains of IRTKS were visualized by fluorescence microscopy 4–5 hr post-induction of fusion protein expression. Exemplary GFP inclusions are marked by arrows. The percentage of cells containing GFP inclusions was determined visually and is indicated at the bottom right of each image.