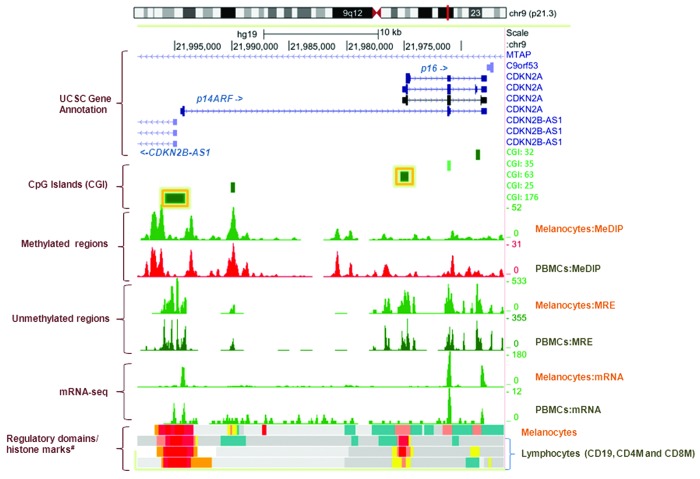
Figure 1. Genome Browser (http://genome.ucsc.edu/) image of the CDKN2A promoter region on human assembly hg19 based on NIH Epigenomics Roadmap data.10 The promoter CpG islands (CGIs) of p16/CDKN2A (CGI:63) and p14ARF/CDKN2A (CGI: 176) analyzed in this study are highlighted with yellow boxes. The CGIs (CGI: 176, 63, 35 and 32) and/or differentially methylated regions technically validated using the DMH-array based method7 in CDKN2A-negative families are annotated to the right of the figure. MeDIP, methylated DNA immunoprecipitation; MRE, methylation-sensitive restrictive enzyme; Melanocytes, normal primary penile foreskin melanocytes (UCSF-UBC-USC and UCSF-UBC); PBMCs, peripheral blood mononuclear cells (UCSF-UBC-UCD and UCSF UBC); lymphocytes, CD19, CD4, and CD8 cells (NIH Epigenomics Roadmap data). Regulatory domains (chromatin state segmentation using a hidden Markov Model [ChromHMM]) and core histone marks: red, active transcriptional start site (TSS); dark salmon, poised TSS; crimson, flanking TSS; orange, active to weak enhancer; yellow, poised enhancer; cadet blue, H3K9me3_K27me3.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.