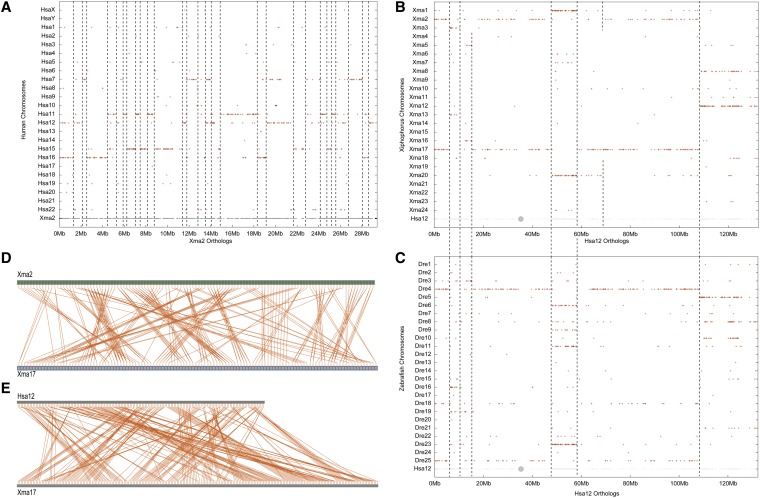
Figure 7.
Conserved syntenies reveal many translocations between platyfish and human genomes. (A) The orthologs of Xma2 reside mainly on five human chromosomes, revealing several translocations and many inversions or transpositions since their last common ancestor. (B) The orthologs of various regions of human chromosome Hsa12 reside on two platyfish chromosomes, as expected from the teleost genome duplication. Because most duplicated regions are coterminus, many translocations occurred before the teleost genome duplication. (C) Comparison of Hsa12 to the zebrafish genome identifies the same duplicated chromosome segments as found in platyfish (B), supporting the origin of many chromosome rearrangements before the divergence of zebrafish and platyfish lineages. (D) Comparison of the orders of paralogous genes on Xma2 and Xma17, which are paralogous chromosomes according to B, identifies many intrachromosomal rearrangements (transpositions and inversions) that occurred after the teleost genome duplication. (E) Comparison of ortholog orders along Xma17 and Hsa12 reveals a substantial number of transpositions and inversions in the 450 MY since their last common ancestor.