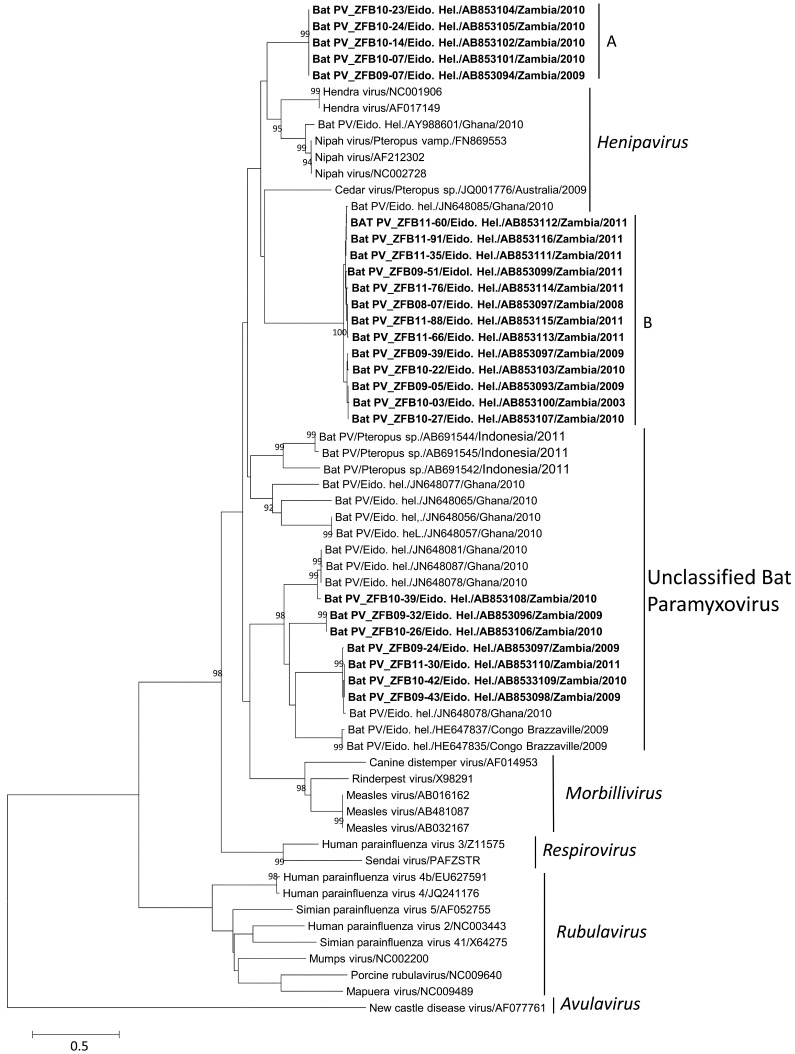
Fig. 1.
Phylogenetic analysis of paramyxovirus samples based on a 530 nucleotide sequence of the polymerase L gene from Zambia (in bold) and other areas of the world. The neighbor joining method was used to construct the phylogram using a confidence level of 1,000 bootstrap replicates. Species names and accession numbers are used to identify both the Zambian samples and the reference isolates. Bootstrap values for 1,000 replicates are indicated as percentage (>90%) and nucleotide substitutions per site as scale bars.