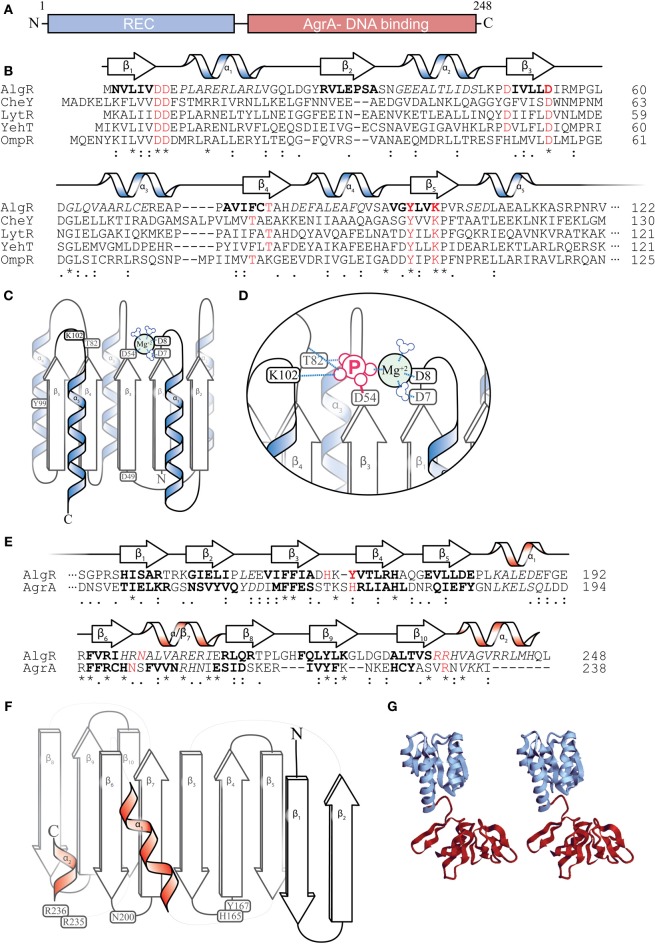
Figure 4.
AlgR Structural predictions. (A) Diagram depicting two major domains of the 249 amino acid AlgR polypeptide. (B) A ClustalW sequence alignment of the amino acid sequence of the N-terminus CheY-like receiver (REC) domain of AlgR and its orthologs. “*” = identical amino acids, “:” = conserved substitutions, “.” = semi-conserved substitutions. Red color denotes select highly conserved amino acids, most with functional importance for phosphorylation, italics indicate alpha helices, bold indicates beta sheets. The predicted secondary structure of AlgR according to Phyre2 is located above the sequence alignment. α = alpha helix, β = beta sheet. (C) An illustrated model depicting the location of conserved amino acids on the secondary structures of the REC domain of AlgR. Aspartate 7, 8, and 54 of CheY-like domains coordinate a magnesium ion, and are required for aspartyl-phosphate transfer from histidine kinase. (D) Once phosphorylated, residues threonine 82 and lysine 102 interact with phosphate ion, changing the conformation of the REC domain. (E) A ClustalW sequence alignment of AlgR and AgrA, with same annotation as (B). Red color denotes select conserved amino acids that are required for DNA interaction in AgrA, and may be required for AlgR interaction with DNA. (F) An illustrated model depicting the location of conserved amino acids on the secondary structures of the LytR/AgrA DNA-binding domain of AlgR. (G) a stereoscopic view of the full AlgR protein modeled, using CheY structure and S. aureus AgrA DNA binding domain.