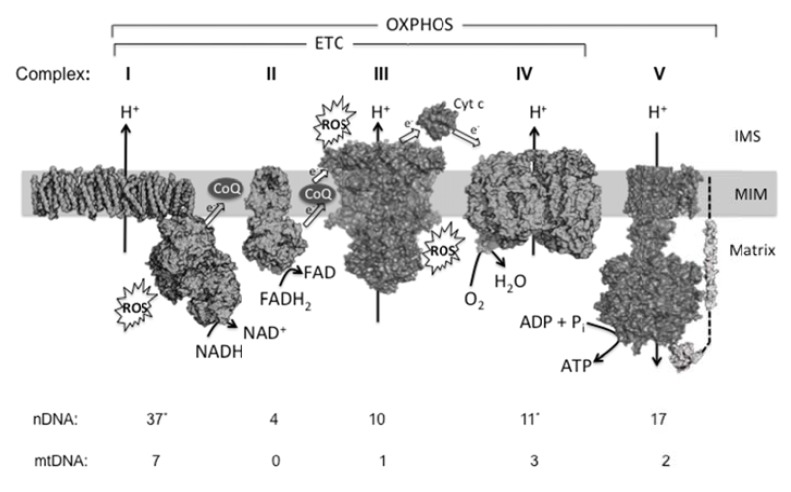
Fig. (2).
Overview of mitochondrial oxidative phosphorylation. The mitochondrial OXPHOS system consists of five multisubunit complexes (I-V) that reside in the mitochondrial inner membrane (MIM). The subunits of these complexes are encoded by the mitochondrial (mtDNA) and nuclear DNA (nDNA). The number of polypeptides being encoded by each genome in mammals is indicated at the bottom. Although organisms other than mammals may exhibit a different pattern of subunits, almost all aerobic organisms carry out OXPHOS according the pathway sketched in this figure. In mammals, complex I is the largest OXPHOS enzyme proposed to consist of 44 different subunits. A simpler bacterial complex I is shown here (3m9s), which is composed of 14 protein chains (corresponding to the “core subunits” from mammals). (*) Recent evidence suggests that the “supernumerary” subunit NDUFA4 hitherto classified as a complex I constituent appears to be a component of complex IV [158], which in this case would have 11 nDNA-encoded subunits. At complex I and complex II (3aee), NADH and FADH2 are oxidized, respectively, and the released electrons are transported to complex III (1be3) via Coenzyme Q10 (CoQ, ubiquinone/ubiquinol). From thereon, electrons are transported to complex IV (2occ) via cytochrome c (Cyt c, 1hrc) and donate to oxygen. Together, complexes I-IV constitute the electron transport chain (ETC). The energy derived from the electron transport is used to expel protons from the mitochondrial matrix across the MIM. This establishes an electrochemical proton-motive force. The backflow of protons is used by complex V (1c17, 1e79) to drive the phosphorylation of ADP and produce ATP. Complexes I and III are thought to be the main ROS production sites.