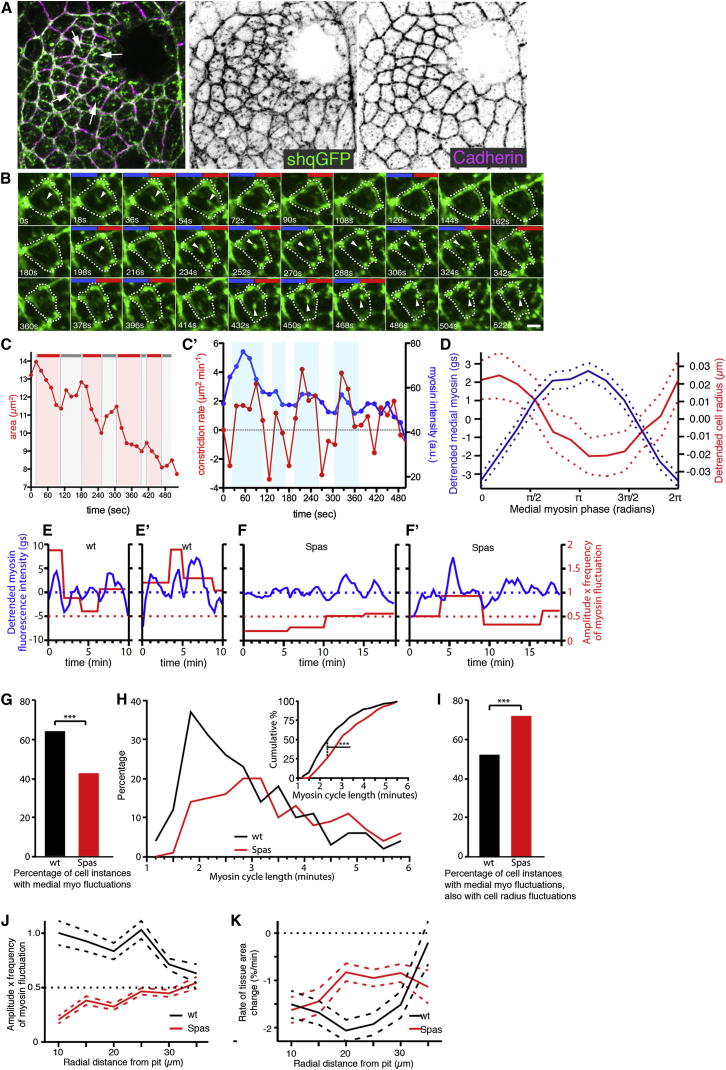
Figure 5.
An Apical Medial Actomyosin Network Involved in Apical Constriction during Tubulogenesis
(A) Myosin II (sqhGFP, green) is organized into an apical junctional and apical medial network across the salivary gland placode (Röper, 2012). The junctional myosin colocalizes with cadherin (magenta), and the medial myosin forms a network-like arrangement across many neighboring cells (arrows).
(B) Still frames of a time-lapse movie of a sqhAX3; sqh::sqhGFP42 embryo (see Figure S4A; Movie S2). The still frames show the fluctuations of medial myosin and the position of the cell cortex (dotted lines) of an exemplary placodal cell; arrowheads point to dynamic, pulsatile concentrations of myosin; blue bars indicate increased myosin II; red bars indicate increased constriction (see C′). The scale bar represents 2 μm.
(C) Apical cell area (μm2) decreases in discrete pulses (red bars) followed by a period of relaxation and stabilization (gray bars).
(C′) Quantification of the constriction rate (μm2 min−1; red) in comparison to medial myosin II intensity (blue) for a single exemplary cell in a placode. An increase in medial myosin II intensity is closely correlated with an increase in constriction rate.
(D) Average medial myosin fluorescence (with trends removed; blue line; gs, grayscale) and cell radius (with trends removed; red line) plotted against phase of medial myosin fluctuation cycle. Two hundred and twelve full cycles of myosin (trough to trough) were pooled and averaged from nine wild-type embryo movies. Dotted lines show 95% confidence intervals.
(E and F) Myosin fluorescence intensity (with trends removed, blue lines) and strength of myosin fluctuation (expressed as amplitude × frequency; red lines) for sample cells. Dotted red lines show the threshold value above which the strength of myosin activity was defined as being periodic.
(E and E′) Two sample wild-type cells (WT).
(F and F′) Two sample MT-depleted cells (Spas). Longer traces are shown for MT-depleted cells because of their longer cycle lengths.
(G–K) Comparison of the average behavior of nine control (wild-type; black) and three MT-depleted embryos (Spas; red). For control embryo data, the number of tracked cell instances (see text) for which it could be established whether a cell was fluctuating or not was 2,877, of which 1,849 exhibited myosin fluctuations. Of the latter, apical radius fluctuated in 929. For MT-depleted embryo data, the number of cell instances was 3,711, 1,584, and 1,106, respectively. See Table S1 for details of statistical analysis.
(G) Percentage of tracked placode cell instances for which medial myosin fluctuations could be detected (see also Movies S3 and S4). ∗∗∗p << 0.0001 using G test of independence.
(H) Distribution of cycle lengths of cells showing myosin fluctuations. Inset: cumulative histograms indicating that cycle lengths of cells still showing fluctuations upon MT depletion were significantly increased. ∗∗∗p << 0.0001 using Kolmogorov-Smirnov test.
(I) Percentage of cell instances with medial myosin fluctuations for which cell-radius fluctuations could also be detected. ∗∗∗p << 0.0001 using G test of independence.
(J) Average strength of myosin fluctuation versus radial coordinate relative to the pit center. Dashed lines are 95% confidence intervals for pooled cell data. Dotted line at 0.5 amplitude × frequency marks the threshold below which cells were not considered to be periodic.
(K) Average rate of change of tissue area versus radial coordinate relative to the pit center (same data as shown in Figures S4E and S4F). Dashed lines show respective 95% confidence intervals.
See also Figures S4 and S5 and Movies S2, S4, and S5.