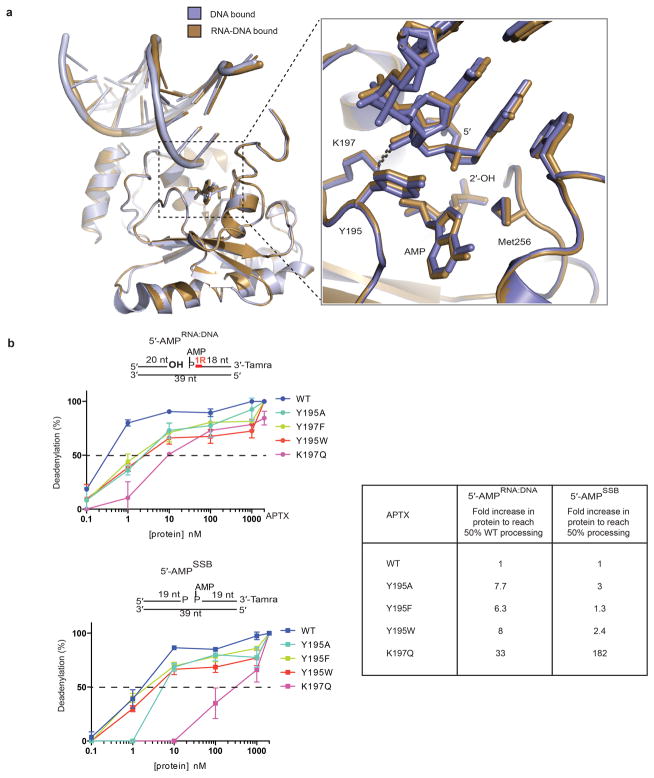
Extended Data Figure 6. Recognition of RNA-DNA and DNA by hAptx.
a, Structural overlays of DNA bound (blue-grey) and RNA-DNA bound (brown) hAptx complex structures. Inset: Interactions at the 5′ terminus reveal similar modes of engagement of RNA-DNA and DNA bound substrates. The β2-β3 loop residues Y195 and K197 orient the 5′ terminus for catalysis. b, Effects of β2-β3 loop mutants on Aptx deadenylation activity. Ten-fold dilutions of Aptx mutant proteins were tested for DNA adenylation activity on 5′-AMPRNA-DNA or 5′-AMPSSB substrates. Mean ± s.d. (n=2 technical replicates) is displayed. Fold increase to of protein to reach 50% activity is relative to the wild type hAptx for each substrate.