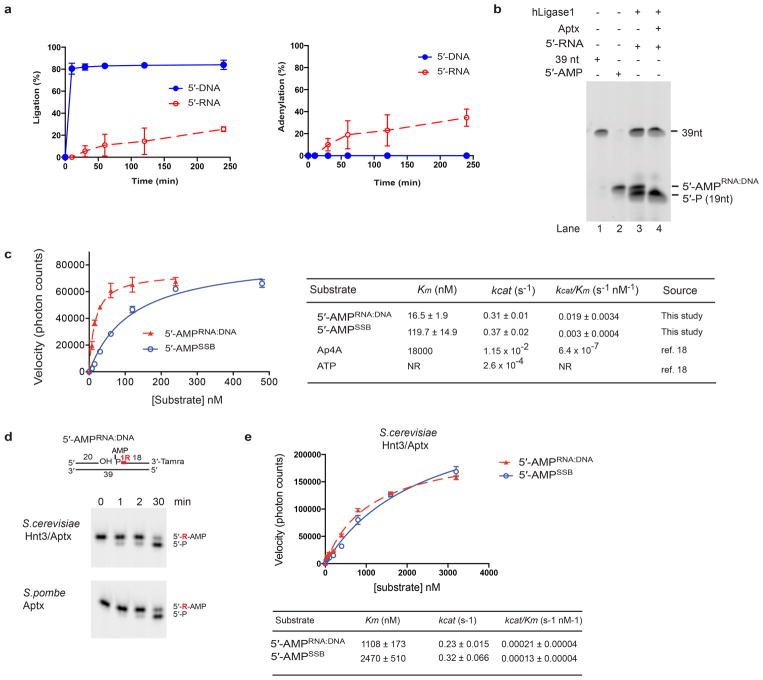
Extended Data Figure 1. Aptx homologs efficiently process products of abortive DNA ligation at RNA-DNA junctions.
a, Abortive DNA ligation reaction time course. DNA ligation reactions for human DNA ligase I were monitored at the indicated time points. The percent ligation was measured by quantitating the total amount of 39 nt ligation product evolved as a percentage of total unprocessed substrate (Fig. 1b, 5′-P species). The percent adenylation was measured by quantitating the total amount of 5′-AMP product evolved as a percentage of total unprocessed substrate. Substrates used are those described in Figure 1b. Mean ± s.d. (n=2 technical replicates) is displayed. b, Recombinant human Aptx specifically processes the adenylated product of DNA ligase I abortive ligation at an incised RNA-DNA junction. DNA ligation reactions (see Fig. 1b) on 5′-RNA substrates were stopped with addition of 5 mM EDTA at 240 min. Recombinant hAptx (165–342) catalytic domain was added following abortive ligation (lane 4). hAptx specifically processed the “5′-AMPRNA:DNA” species generated by abortive ligation at the RNA-DNA junction, but does not process the 19 nt 5′-P substrate, or the 39 nt ligation product, confirming the “5′-AMPRNA:DNA” species is a bona fide adenylation product. c, Kinetic parameters of hAptx processing of oxidative DNA damage and RNA-DNA junction derived adenylated substrates. Mean ± s.e.m (n=2 technical replicates) is displayed. d, 5′-AMPRNA-DNA deadenylation activity is an evolutionary conserved function of fission and budding yeast Aptx/Hnt3. Reaction conditions used were as for Fig. 1d. e, Kinetic parameters of S. cerevisiae Aptx (Hnt3) deadenylation processing. Kinetic experiments were performed as described in the Methods. Mean ± s.e.m (n=2 technical replicates) is displayed.