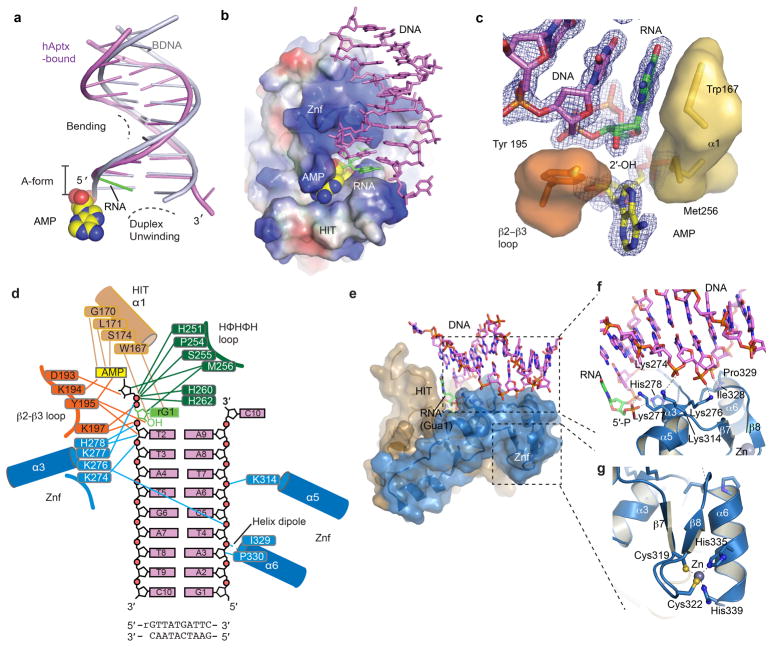
Extended Data Figure 5. hAptx RNA-DNA interactions.
a, Structural distortions of the hAptx bound RNA-DNA. A cartoon representation of the duplex from the hAptx/RNA-DNA/AMP/Zn reaction product complex (magenta and green) is shown superimposed on an ideal B-form DNA duplex (gray), showing distortion of the 5′-terminal nucleotides from B-form geometry. b, Electrostatic potential representation of the hAptx HIT-Znf DNA interaction interface is displayed with electropositive (blue) electronegative (red) and hydrophobic (white) surfaces. An extended positively charged surface of the Znf mediates sequence non-specific RNA-DNA contacts. Hydrophobic base stacking stabilizes the exposed ribonucleotide base. c, Trp167 and Tyr195 anchor the terminal ribonucleotide (green), and envelop the adenylate lesion (yellow). An omit σ-A weighted 1.95 Å Fo-Fc map is displayed contoured at 3.0 σ overlaid upon the AMP and RNA-DNA duplex product complex. The RNA-DNA duplex and AMP were excluded from the model for electron density map calculation. d, hAptx protein RNA-DNA and AMP lesion binding contacts are displayed schematically. e, The HIT (tan) and Znf (blue) surfaces mold a contiguous RNA-DNA damage interacting surface. f, Molecular details of the Znf structure-specific DNA damage binding interface. DNA-protein contacts are mediated by four basic side chains (Lys 276, Lys277, His 278, Lys314) of Znf helicies α3 and α5. Additional sugar-phosphate backbone contacts from Pro 329 and Ile 328, as well as the electropositive helix-dipole of helix α6 also engage the undamaged strand. g, Zn-binding by the hAptx C2H2 zinc finger subdomain. The Zn is coordinated with tetrahedral geometry by four zinc binding residues (Cys319, Cys322, His335 and His339) and scaffolds folding of the Znf domain.