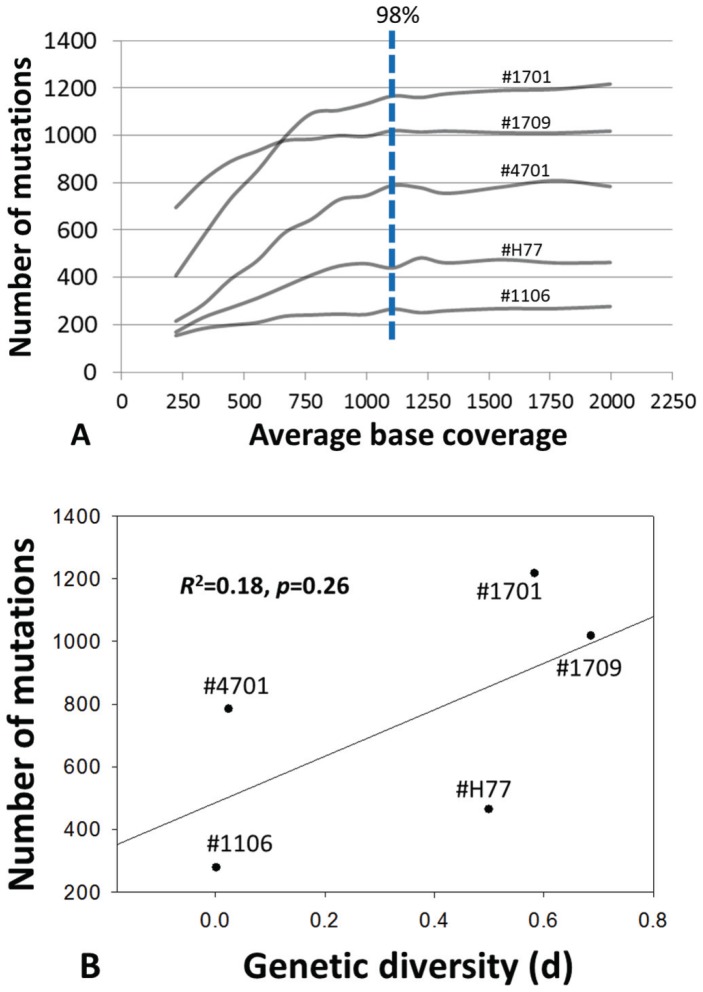
Figure 1. Simulation of the sequencing depth required for stable quantitation of HCV mutation load at a resolution of 1% mutation frequency in population.
Five patient samples with known intra-population genetic diversity were subject to 454 sequencing. Starting at an average base coverage of 1100x, the genome-wide HCV mutation load became stable with overall 98% saturation (A) and had a weak correlation with genetic diversity measured by cloning and sanger sequencing (R 2 = 0.18, p = 0.26) (B).