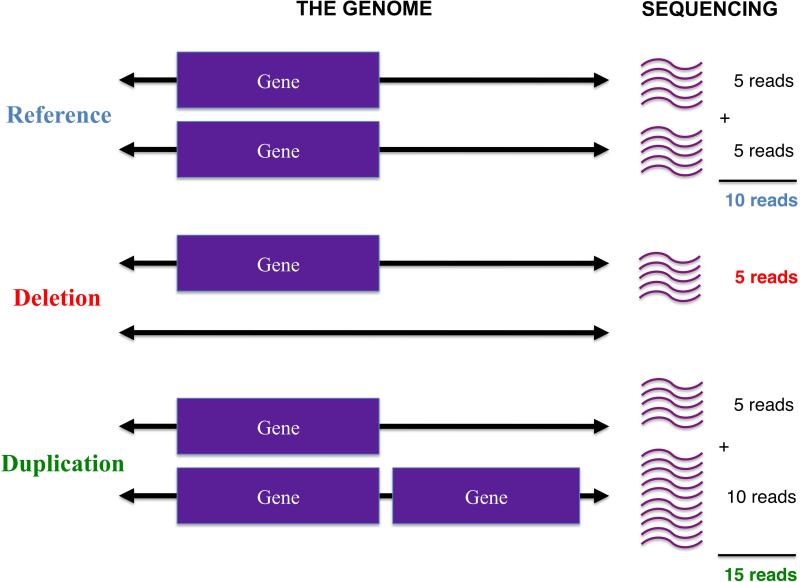
Figure 1. How genomic copy number affects depth of sequencing.
Depicted are a reference individual with two copies of a gene, an individual with only a single copy (deletion of gene on one chromosome), and an individual with an extra copy of a gene in their genome (duplication). In an idealized setting where each individual's genome is targeted at an average of 10× coverage, 5 of those reads will come from the maternal chromosome and 5 from the paternal. If one of those chromosomes is missing that gene (deletion), then only 5 reads for that gene will be observed; on the other hand, if an individual has a duplication of that gene, then a total of 15 reads will be found. In reality, noise and biases in the data make it difficult to easily and directly read off genomic copy number from such coverage information. The purpose of XHMM is to automate a procedure for performing such inference in a robust manner.