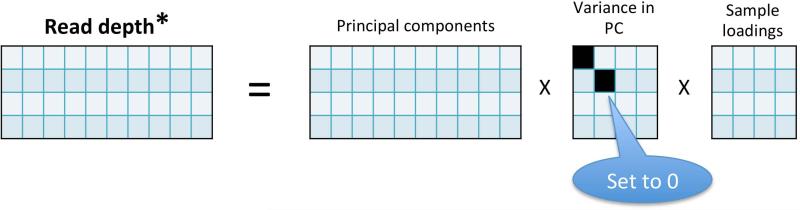
Figure 3. Normalization by removal of top principal components.
In this “toy” example (of 4 samples targeted at 10 exons), the 4×10 read depth matrix is first decomposed into the principal components (the key axes in which the read depth varies), the variance of the data in each such component, and the sample loadings (the coefficients of each sample in each component). Then, in this example, it is estimated (not shown) that the two largest principal components correspond to non-CNV read depth effects, based on their large relative contribution to the variance of the read depth data. These components are thus removed by zeroing them out, and the reconstructed read depth matrix will be used for CNV calling.