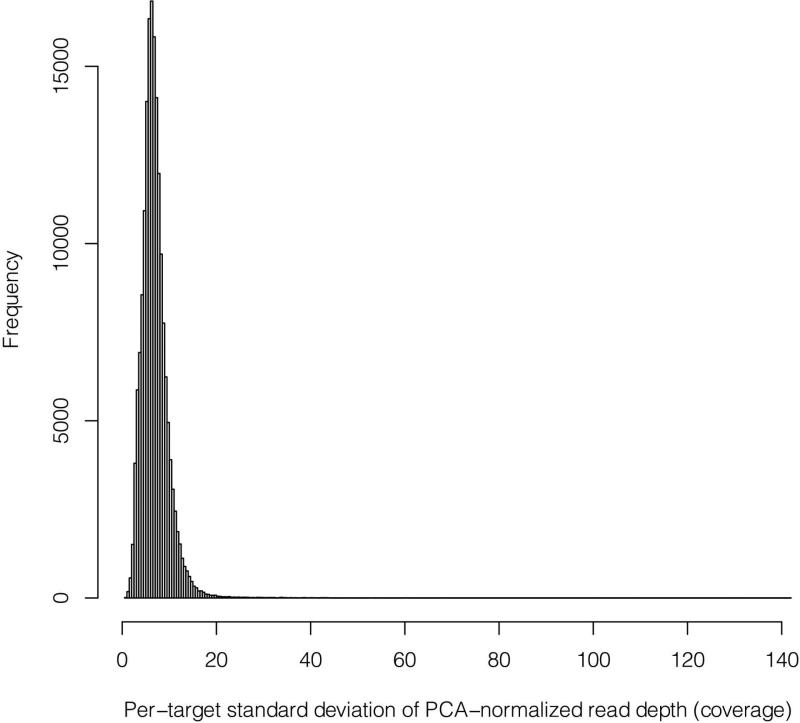
Figure 9. Distribution of post-normalization target variance.
Before XHMM calculates z-scores and the HMM is run to call CNV for each sample (CNV “discovery”), we perform a final filtering step. Specifically, we remove any targets that have “very scattered” read depth distributions post normalization. These can be thought of as targets for which the normalization may have failed, and it is better to remove such strong effects (still likely to be artifacts) to prevent them from drowning out other more subtle signals. In detail, we removed any targets with large standard deviations of their post-normalization read depths across all samples. As a (proto-typical) example, we see here that the small fraction of targets with standard deviations any larger than the 30 to 50 range were removed (in this case, for a scenario of ~100× mean sequencing coverage).