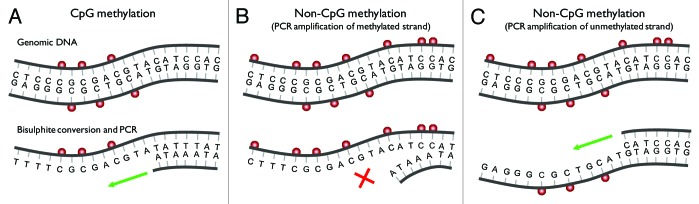
Figure 1. Bisulphite PCR can cause underestimation of the levels of non-CpG methylation. (A) A short region of genomic DNA with symmetrical CpG methylation (red spheres). Bisulphite converts cytosine to uracil (recognized as thymine during PCR), whereas 5-methylcytosine remains non-reactive. The sequence is then amplified (green arrow) using primers specific to the converted sequence. Primers are usually designed assuming that all cytosine in a non-CpG dinucleotide context are converted to uracil (and hence unmethylated). (B) The same region of DNA with additional methylation at non-CpG sites. Methylation at non-CpG dinucleotides within the binding site of the primer will result in protection from bisulphite conversion and loss of primer complementarity. Under these circumstances all molecules containing non-CpG methylation fail to amplify (red cross) and are not detected. (C) Non-CpG methylation, which is inherently asymmetrical, will remain undetected following PCR amplification (green arrow) using primers specific to the unmethylated strand.
