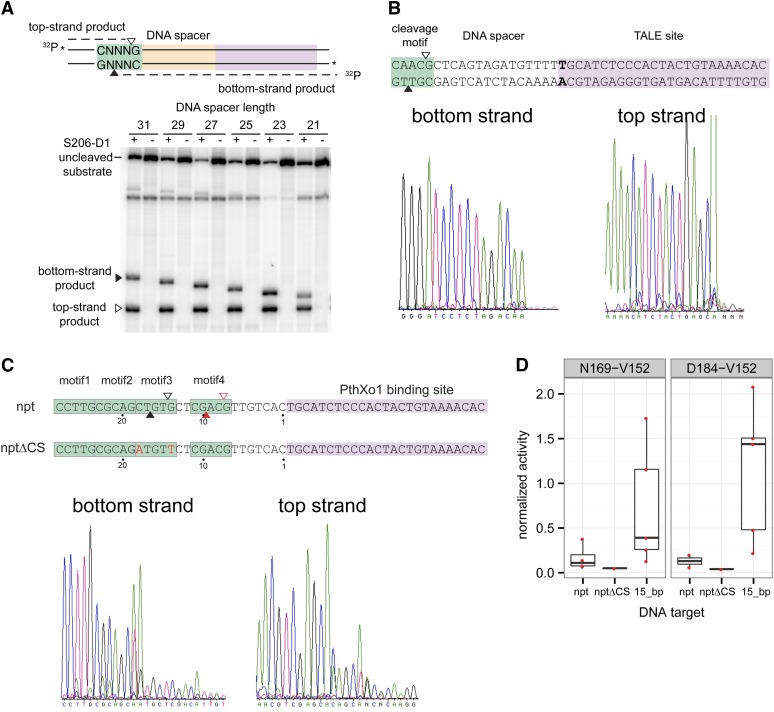
Figure 2.
Mapping of Tev-mTALEN cleavage sites. (A) Shown at the top is a schematic of double-strand oligonucleotide substrate labeled on top-strands and bottom-strands. The top-strand nicking product is indicated by an open triangle (▵), and the bottom-strand nicking product is indicated by a filled triangle (▴). Below is a representative denaturing polyacrylamide gel of cleavage reactions with the S206-T120 Tev-mTALEN. Top-strand and bottom-strand products are represented by open (▵) and filled (▴) triangles, respectively. (B) In vitro mapping of N169-T120 Tev-mTALEN cleavage sites on supercoiled plasmid substrates containing a 15-bp spacer. Representative ABI traces of run-off sequencing reactions to determine the cleavage sites (taking into account that an extra "A" is added during the sequencing reactions). The cleavage sites are mapped to the complement of the sequencing trace shown. (C) In vitro cleavage mapping on nptII-PthXo1 plasmid substrates that contain four CNNNG motifs. The open (▵) and filled (▴) red triangles indicate secondary cleavage sites inferred from run-off sequencing. The electropherograms shown are derived from the nptII substrate. (D) In vivo activity of the N169-V152 and D184-V152 Tev-mTALENs on nptII and nptIIΔCS substrates (see text) measured in the yeast-based lacZ repair assay. Activity is normalized to a homodimeric ZFN control and is shown relative to activity on the wild-type I-TevI target with 15-bp spacer.