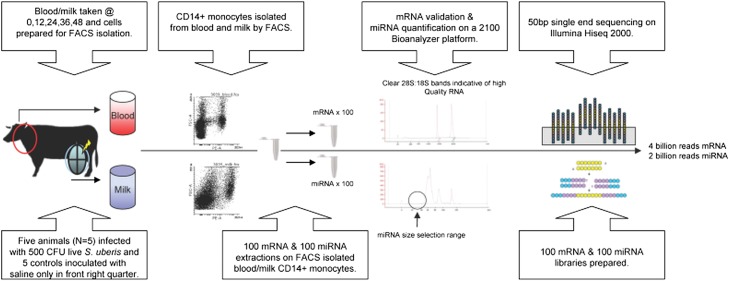
Figure 1.
Overview of the experimental design. Five Holstein Friesian animals were infected via the teat canal of the right front quarter with approximately 500 CFU of a mastitis-causing pathogen, Streptococcus uberis 0140, in 10 ml saline. Five control animals were inoculated with saline only. Milk and blood samples were obtained from each animal at 0, 12, 24, 36, and 48 hr after infection (or mock infection). Milk-derived and blood-derived CD14+ monocytes were isolated by fluorescence-activated cell sorting (FACS) from each sample. mRNA and miRNA were extracted and 200 Illumina-compatible libraries were prepared for sequencing on a Hiseq 2000 machine. More than four billion mRNA reads and more than two billion miRNA reads were sequenced in total.