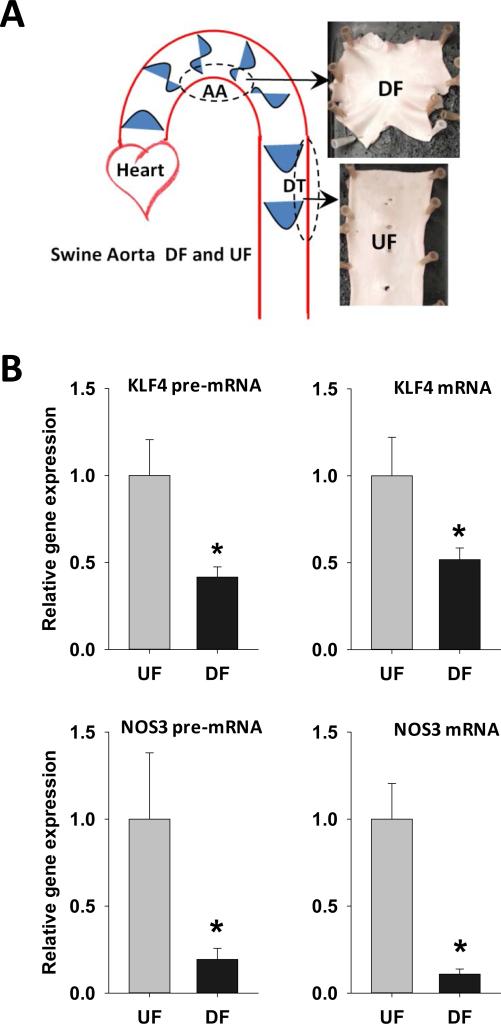
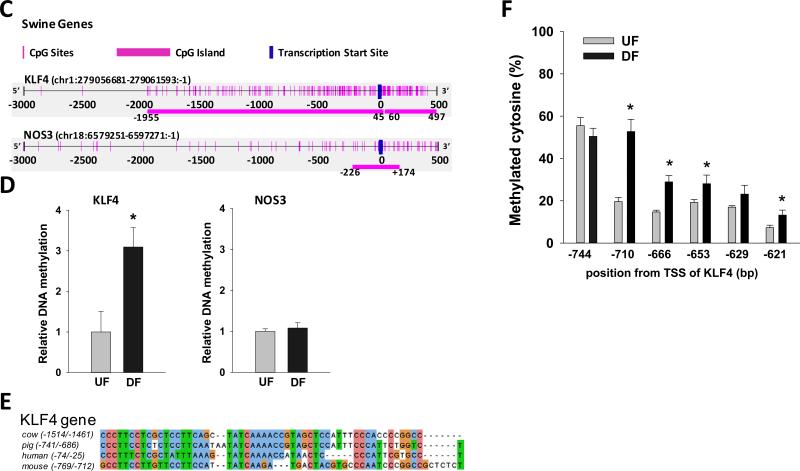
Figure 6. Differential gene expression and promoter methylation of swine arterial endothelial KLF4and NOS3 in vivo.
(A) Schematic illustration of targeted UF and DF regions in swine aorta. Endothelial cells were scraped gently from the descending thoracic aorta (DT) where undisturbed flow (UF) is dominant, and from the inner curvature of aortic arch (AA) where disturbed flow (DF) is dominant. (B) mRNA and pre-mRNA of swine KLF4 and NOS3 were determined by qPCR. Data are normalized to the geometric mean of GAPDH and PECAM1 and are expressed as mean ± SEM fold of UF. (C) Schematic illustration of the CpG island and CpG sites in swine KLF4 and NOS3 promoters. (D) Methylation of KLF4 promoter and NOS3 promoter in UF and DF region of swine aorta was determined by using MSP targeting CpG-rich region. Data are normalized to UBB promoter without CpG sites and expressed as mean ± SEM fold of UF. (E) Alignment of KLF4 promoter region in multiple mammalian species showing a highly conserved region of MEF2 binding sequence. (F) Methylation level of individual CpG sites in swine KLF4 promoter determined by bisulfite-pyrosequencing. *Different from UF. P < 0.05. n= 6.