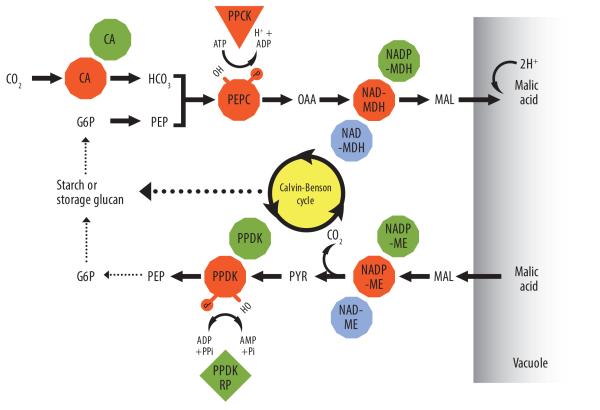
Figure 4.
Proposed model of a minimum gene set encoding enzymes of core CAM carboxylation–decarboxylation modules and their known regulation based upon enzyme activity [76, 141, 142] and mRNA expression studies [58, 59, 81]. Key metabolites include: G6P, glucose-6-phosphate; PEP, phosphoenolpyruvate; OAA, oxaolacetate; MAL, malate; PYR, pyruvate. Broken arrows indicate multiple metabolic steps. Key enzymes include: CA, carbonic anhydrase; PEPC, phosphoenolpyruvate carboxylase; PPCK, PEPC kinase; NAD(P)-MDH, NAD or NADP-dependent malate dehydrogenase; NAD(P)-ME, NAD- or NADP-dependent malic enzyme; PPDK, pyruvate orthophosphate dikinase; PPDK-RP, PPDK regulatory protein. The involvement of PPDK-RP is inferred from studies in C3 and C4 species [91]. Colors indicate subcellular locations of enzymes: cytosol (orange), chloroplasts (green), and mitochondria (blue).