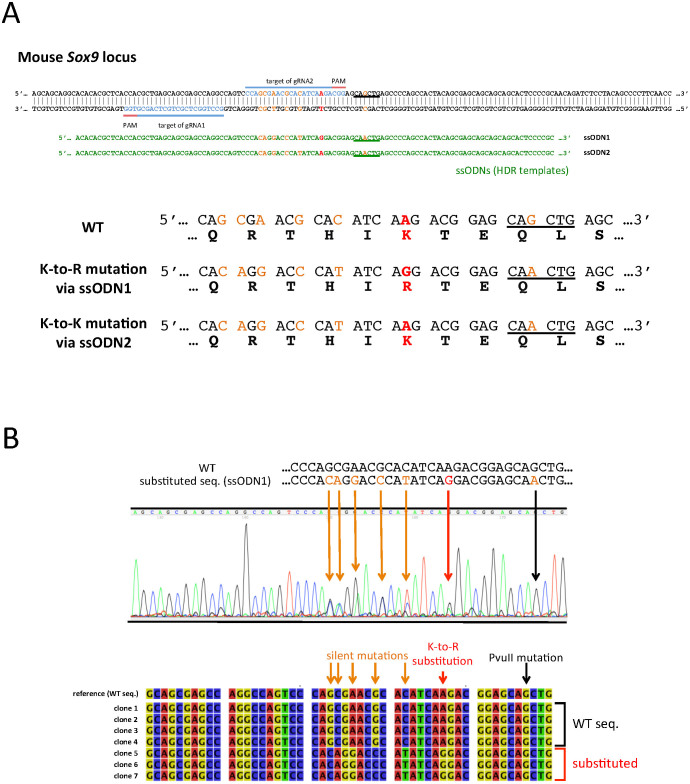
Figure 2. Introduction of the K396R substitution in Sox9 locus.
(A) A schematic illustration demonstrating the locations of the gRNAs and ssODNs along with the mouse Sox9 locus. The blue bars and letters indicate the positions of gRNA targets, while red bars highlight the PAM sequences. The sequences of ssODNs are shown in green letters at the corresponding positions to the Sox9 locus. Red letters indicate the substitution target site in the Sox9 locus and the corresponding mismatched nucleotides in the ssODNs. The deduced amino acid sequences from wild type (WT) and substituted sequences are shown at the bottom, with red letters indicating the target and the results of the substitution. Orange letters in the sequences are the targets of silent mutations, which do not cause amino acid conversion. The PvuII site is underlined in the Sox9 loci which had mutated in ssODNs. (B) The results of the sequence analysis of the Sox9 locus of a representative mouse, which had a monoallelic K-to-R substitution. The Sox9 locus around the gRNA target site was PCR amplified and directly sequenced (Upper panel), or was cloned into the plasmid and sequenced independently (Lower panel). The upper panel shows the electropherogram of the direct sequencing result of a mouse obtained in experiment 1 shown in Table 2, in which one allele received a K-to-R substitution. The WT and donor DNA sequences are shown on the top, with orange and red letters indicating mutated nucleotides. The orange and red arrows indicate the overlapping peaks caused by monoallelic substitutions, and a black arrow indicates the non-overlapping peak despite the fact that the ssODN (donor DNA) had a mutated nucleotide. The lower panel shows the sequence alignment of seven independent clones. The seven sequences were separated into two types: the WT and substituted sequences.