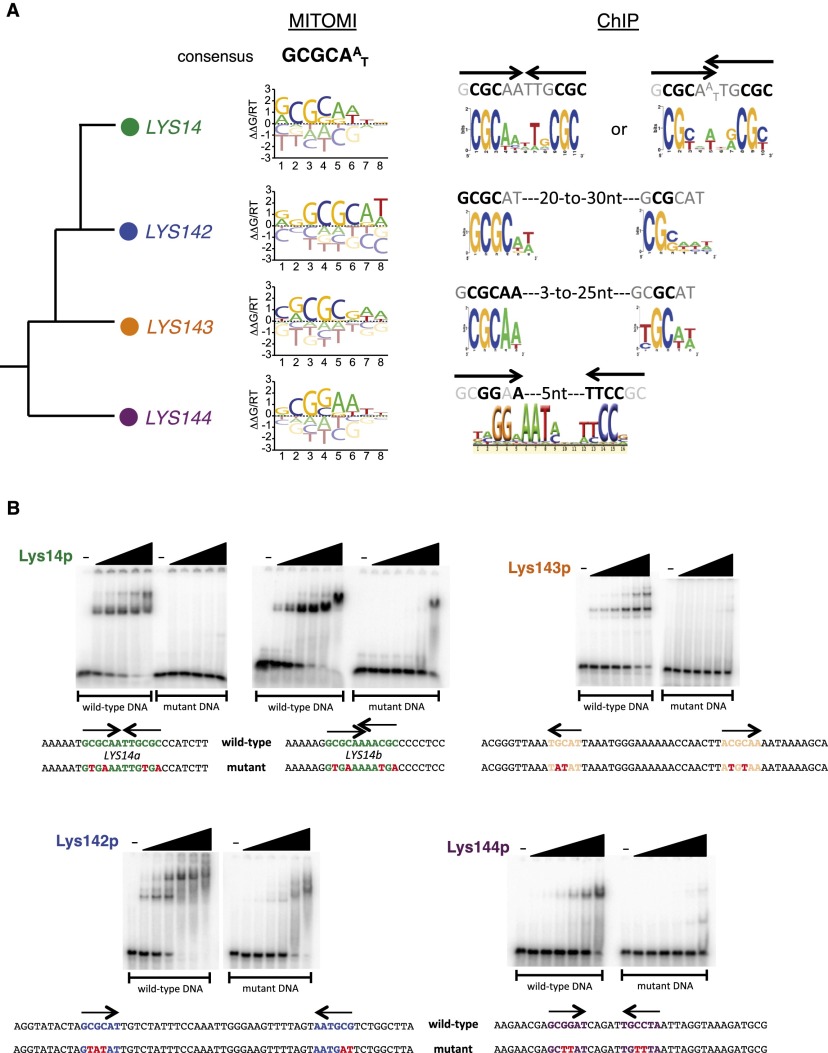
Figure 2.
DNA motifs preferred by C. albicans LYS transcription regulators. (A) DNA motifs derived from MITOMI and ChIP data sets. (B) Gel shift assays showing binding of the Lys proteins to their predicted binding sites. 32P-labeled DNA fragments (∼0.4 nM) containing the predicted wild-type or mutant LYS-binding sites were incubated with increasing concentrations of purified Lys protein (0, 0.039, 0.156, 0.625, 2.5, 10, and 40 nM) for 30 min at room temperature in standard EMSA buffer and resolved in 6% polyacrylamide gels run with 0.5× TGE. Point mutations introduced in the binding sites are indicated in red.