Significance
Spermatogonial stem cells (SSCs) are at the foundation of spermatogenesis. Previous studies on SSC self-renewal have focused on the positive regulation, but we hypothesized that negative regulation of self-renewal is equally important in understanding SSC self-renewal. We show here that F-box and WD-40 domain protein 7 (FBXW7), a component of the Skp1-Cullin-F-box–type ubiquitin ligase, negatively regulates SSC self-renewal. FBXW7 is expressed only in the undifferentiated spermatogonia compartment, and its overexpression compromises SSC activity whereas Fbxw7 deficiency enhances SSC colonization and caused accumulation of undifferentiated spermatogonia. We also demonstrate that FBXW7 regulates stability of myelocytomatosis oncogene (MYC), which increased SSC activity upon overexpression. These results suggest that FBXW7 counteracts with positive regulators of self-renewal by degrading MYC and demonstrate the importance of negative regulators of self-renewal in understanding SSC homeostasis.
Abstract
Spermatogonial stem cells (SSCs) undergo self-renewal divisions to support spermatogenesis throughout life. Although several positive regulators of SSC self-renewal have been discovered, little is known about the negative regulators. Here, we report that F-box and WD-40 domain protein 7 (FBXW7), a component of the Skp1-Cullin-F-box–type ubiquitin ligase, is a negative regulator of SSC self-renewal. FBXW7 is expressed in undifferentiated spermatogonia in a cell cycle-dependent manner. Although peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 (PIN1), essential for spermatogenesis, is thought to destroy FBXW7, Pin1 depletion decreased FBXW7 expression. Spermatogonial transplantation showed that Fbxw7 overexpression compromised SSC activity whereas Fbxw7 deficiency enhanced SSC colonization and caused accumulation of undifferentiated spermatogonia, suggesting that the level of FBXW7 is critical for self-renewal and differentiation. Screening of putative FBXW7 targets revealed that Fbxw7 deficiency up-regulated myelocytomatosis oncogene (MYC) and cyclin E1 (CCNE1). Although depletion of Myc/Mycn or Ccne1/Ccne2 compromised SSC activity, overexpression of Myc, but not Ccne1, increased colonization of SSCs. These results suggest that FBXW7 regulates SSC self-renewal in a negative manner by degradation of MYC.
Spermatogonial stem cells (SSCs) provide the foundation for spermatogenesis by undergoing self-renewing division (1, 2). SSCs reside in a special niche microenvironment where they are provided with self-renewal factors that stimulate proliferation. Glial cell line-derived neurotrophic factor (GDNF) is secreted from the niche to promote SSC self-renewal (3). Studies on SSCs are hampered because SSCs constitute only 0.02–0.03% of germ cells, which amounts to 2–3 × 104 cells per mouse testis (2, 4). Moreover, the lack of a specific marker has made it difficult to distinguish SSCs from committed spermatogonia. In 1994, a germ cell transplantation technique was developed (5), in which transplanted SSCs reinitiate spermatogenesis and produce functional sperm following microinjection into seminiferous tubules of testes. Moreover, the addition of GDNF stimulated self-renewing division of SSCs and allowed long-term expansion in vitro. Cultured cells, designated as germ-line stem (GS) cells, proliferate for at least 2 y and expand to 1085-fold, creating a unique opportunity to collect a large number of SSCs for biochemical and molecular biological analyses (6).
Using these techniques, molecular regulators of SSC self-renewal have been analyzed. It is now considered that GDNF activates Harvey rat sarcoma virus oncogene (HRAS) via Src family kinase molecules, and cells transfected with activated HRAS underwent self-renewal division without exogenous cytokines (7). Chemical inhibition of thymoma viral proto-oncogene (AKT) or mitogen-activated protein kinase kinase 1 (MAP2K1), both of which are downstream molecules of HRAS, abrogated GS cell proliferation (8–10), suggesting that they are necessary for self-renewal division. When active Akt or Map2k1 was overexpressed in GS cells, Akt- or Map2k1-transfected cells could proliferate with only fibroblast growth factor 2 (FGF2) or GDNF, respectively (8, 9). These signals up-regulate Etv5 and Bcl6b (8, 11), which work in combination with other constitutively expressed transcription factors, such as zinc finger and BTB domain containing 16 (Zbtb16) or Taf4b, to drive SSC self-renewal.
Although these studies have identified several positive regulators of SSC self-renewal, little is known about the negative regulators, which would be equally important in understanding stem-cell kinetics. In this study, we analyzed the function of the F-box protein FBXW7 (also known as Fbw7, Cdc4, Fbxw6, or Fbxo30), a substrate recognition subunit of the Skp1-Cullin-F-box complex (12). FBXW7 catalyzes the ubiquitination of various molecules involved in cell-fate decision and cell-cycle regulation. We hypothesized that degradation of these critical regulators by FBXW7 influences cell-cycle regulation or fate commitment in SSCs. We tested this hypothesis by taking advantage of SSC cultures and transplantation techniques.
Results
Restricted Expression of FBXW7 in Spermatogonia.
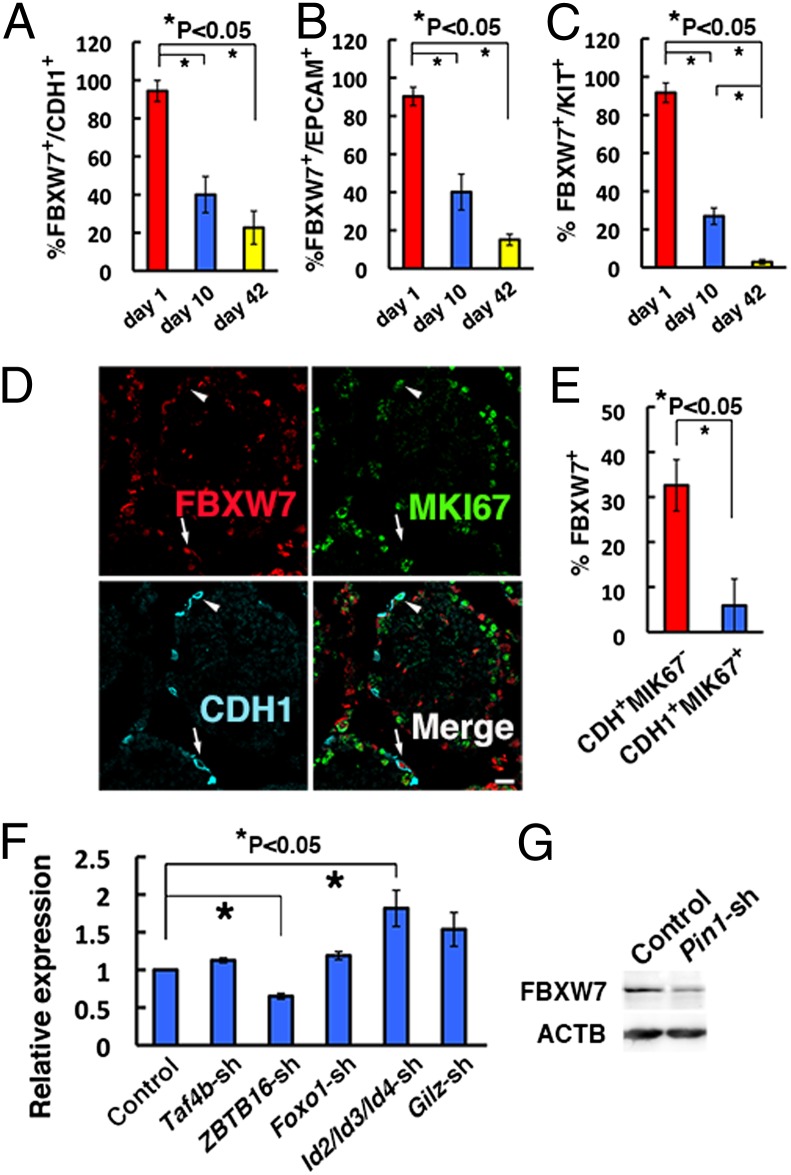
We examined FBXW7 expression in seminiferous tubules of 1-, 10-, and 42-d-old mice (Fig. S1 A–D). In 1-d-old testes, FBXW7 was expressed in most of the gonocytes, which are precursors of spermatogonia. However, FBXW7 expression was significantly decreased in 10-d-old mouse testes, which contain proliferating spermatogonia. All stages of germ cells are present in 42-d-old testes, but FBXW7 was found in only 22.7% of cadherin 1 (CDH1)-expressing undifferentiated spermatogonia and 15.1% of epitherial cell adhesion molecule (EPCAM)-expressing spermatogonia (Fig. 1 A and B and Table S1). FBXW7 was rarely found in kit oncogene (KIT)-expressing differentiating spermatogonia of adult testes (Fig. 1C). These results suggested that FBXW7 is expressed during the primitive stages of spermatogenesis but down-regulated during differentiation. Interestingly, FBXW7 expression was more frequently found in CDH1+ antigen identified by monoclonal antibody Ki67 (MKI67)– spermatogonia compared with CDH1+MKI67+spermatogonia (Fig. 1 D and E), which suggests that FBXW7 is expressed in a cell cycle-specific manner.
Fig. 1.
Expression of FBXW7 in spermatogonia. (A–C) Expression of FBXW7 protein in CDH1+ (A), EPCAM+ (B), or KIT+ (C) cells during postnatal testis development. At least 12, 15, and 14 tubules were counted in 1-, 10-, and 42-d-old testes, respectively. (D) Triple immunohistochemistry of FBXW7, MKI67, and CDH1 in 42-d-old testes. At least 17 tubules were counted. Arrows indicate CDH1+MKI67− cells with FBXW7 expression. Arrowheads indicate CDH1+MKI67+ cells without FBXW7 expression. (E) Proportion of CDH1+ cells with FBXW7 expression. (F) Real-time PCR analysis of Fbxw7 expression following depletion of indicated genes by shRNA (n = 6). (G) Western blot analysis of FBXW7 expression following Pin1 depletion. (Scale bar: D, 20 μm.)
To examine the regulation of Fbxw7, we used GS cells and enriched germ cells from pup testis. Self-renewal factors, including FGF2 and GDNF, did not show an apparent effect (Fig. S2 A and B). GDNF also did not induce apparent changes in pup testis cells after gelatin selection, which are enriched for SSCs (Fig. S2 C and D). Screening of Fbxw7 regulators using lentiviruses expressing short hairpin RNA (shRNA) revealed that depletion of Zbtb16 reduced the expression of Fbxw7 whereas depletion of inhibitor of DNA binding (Id)2/3/4 increased Fbxw7 mRNA expression (Fig. 1F, Fig. S2E, and Table S2). However, Western blotting showed no significant changes after these treatments (Fig. S2F). Nevertheless, because several F-box proteins, including FBXW7, are unstable and regulated posttranslationally (13), we also analyzed the impact of Pin1, a prolyl isomerase that was reported to change FBXW7 protein expression: Pin1 overexpression down-regulated FBXW7 whereas its depletion led to elevated FBXW7 expression in human cancer cells (14). Contrary to our expectation, Pin1 depletion down-regulated FBXW7 expression (Fig. 1G and Fig. S2 E and G). In contrast, Pin1 overexpression did not influence FBXW7 expression (Fig. S2 H and I), suggesting that PIN1 is necessary but not sufficient for maintaining FBXW7 expression.
Overexpression of Fbxw7 Suppresses the Proliferation of SSCs.
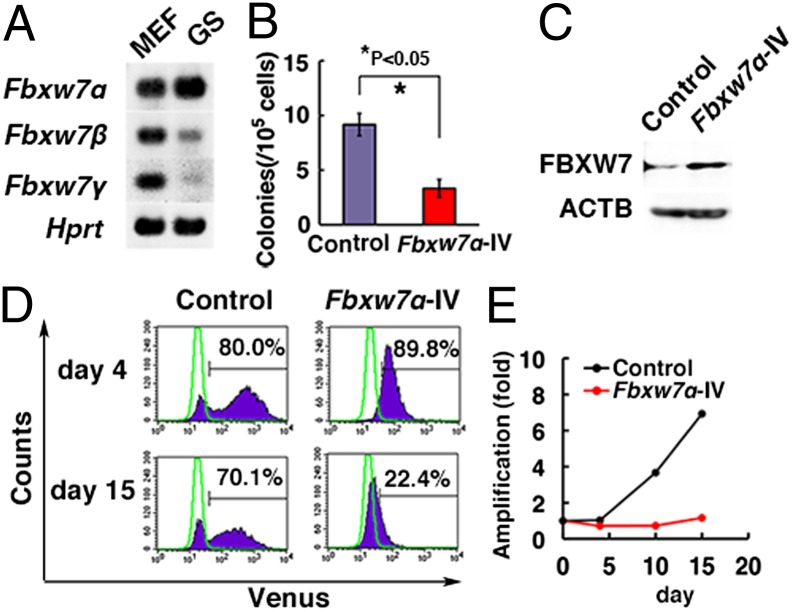
There are three isoforms of Fbxw7 (Fbxw7α, -β, and -γ), and reverse-transcriptase (RT)-PCR analysis showed that Fbxw7α was strongly expressed in GS cells (Fig. 2A). To examine the impact of Fbxw7α overexpression in SSCs, lentivirus-mediated transduction of Fbxw7α and Venus was performed in testis cells from 10-d-old C57BL/6 Tg14(act-EGFP)OsbY01 (green) transgenic mice that ubiquitously express enhanced green fluorescent protein (EGFP). After overnight infection, the viral supernatant was removed, and, after 2 d, Fbxw7α-transduced cells were transplanted into seminiferous tubules of WBB6F1-W/Wv (W) mice. Testis cells transduced with an Fbxw7α-expressing vector produced 3.3 ± 0.8 colonies per 105 cells whereas control testis cells produced 9.2 ± 1.0 colonies per 105 cells (n = 12), and this decrease in colony number was statistically significant (Fig. 2B and Fig. S3A).
Fig. 2.
Decreased SSC activity with Fbxw7 overexpression. (A) RT-PCR analysis of Fbxw7 isoform expression in GS cells and mouse embryonic fibroblasts (MEFs). (B) Colony counts after Fbxw7α overexpression and transplantation of green mouse testis cells. Results of two experiments (n = 12). (C) Western blot analysis of Fbxw7α expression in GS cells transduced with Fbxw7α. Cells were recovered 4 d after transfection. (D) Flow cytometric analysis of GS cells transduced with Fbxw7α. Cells were analyzed at the indicated time points. Green lines indicate GS cells without transfection. (E) Proliferation of GS cells following Fbxw7α overexpression.
To understand the mechanism mediating the decrease in SSC induced by Fbxw7α overexpression, we performed transfection experiments using GS cells from B6-TgR(ROSA26)26Sor (ROSA26) mice. Flow cytometry analysis showed that Fbxw7α overexpression conferred a selective disadvantage for GS cell proliferation (Fig. 2 C and D). Although the proportion of cells displaying Venus fluorescence was comparable 4 d after infection, it dropped significantly after 15 d. Consistent with this observation, cell recovery was also decreased by Fbxw7α transfection, which suggested that Fbxw7α overexpression suppresses GS cell proliferation (Fig. 2E). This growth suppression occurred despite decreased expression of several cyclin-dependent kinase inhibitors (CDKIs) (Fig. S3B). Adding GDNF did not influence colonization levels of primary pup testis cells (Fig. S3C). Taken together, these results suggest that Fbxw7α overexpression has a negative impact on SSC self-renewal.
Conditional Deletion of Fbxw7 Impairs Spermatogenesis.
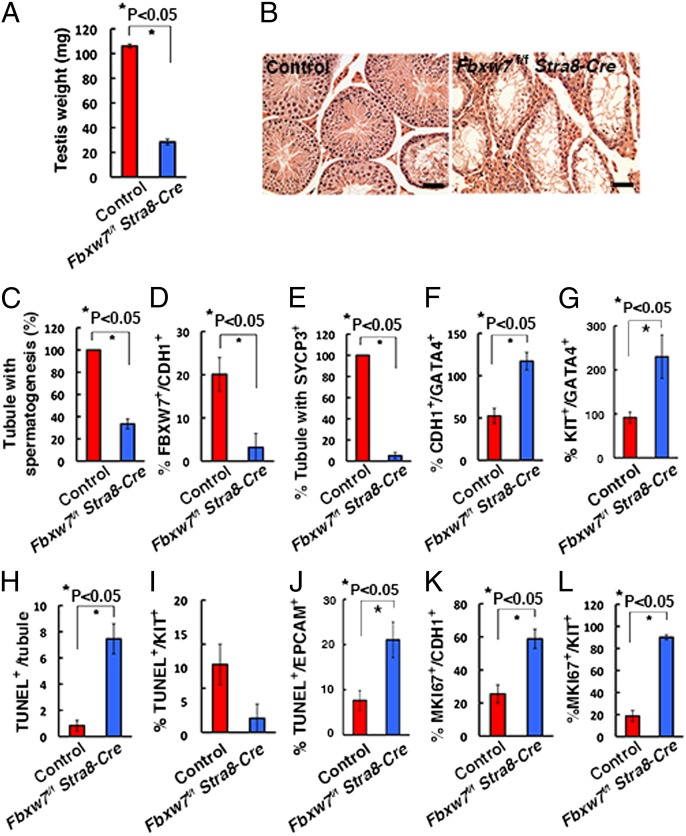
To examine the impact of Fbxw7 deletion on spermatogenesis, we crossed mice homozygous for the floxed Fbxw7 allele (Fbxw7f/f) with stimulated by retinoic acid gene 8 (Stra8)-Cre transgenic mice, which express Cre recombinase under the Stra8-promoter (15). Stra8 is a retinoic acid-responsive gene, which is first expressed a few days after birth in the majority of ZBTB16+ undifferentiated spermatogonia. Testes of Fbxw7f/f Stra8-Cre adult mice were apparently smaller than those of controls (Fbxw7+/+ Stra8-Cre) (Fig. 3A). Histological analysis of 2-mo-old Fbxw7f/f Stra8-Cre testes revealed significantly reduced germ cells in mutant testes, and few meiotic cells were found (Fig. 3B). The number of tubules with spermatogenesis, as defined by the tubules with multiple layers of germ cells, was significantly decreased by Fbxw7 deficiency (Fig. 3C).
Fig. 3.
Impaired spermatogenesis in Fbxw7f/f Stra8-Cre mice. (A) Testis weight of 2-mo-old Fbxw7f/f Stra8-Cre mice (n = 8, control; n = 6, Fbxw7f/f Stra8-Cre). (B) Histological appearance of Fbxw7f/f Stra8-Cre testis. (C) Tubules with spermatogenesis, defined as the presence of multiple layers of germ cells in the entire circumference of the tubules. At least 207 tubules were counted. (D) Expression of FBXW7 in CDH1+ cells. At least 20 tubules were counted. (E) Expression of SYCP3 in KIT+ cells. At least 22 tubules were counted. (F) Ratio of CDH1+ spermatogonia and GATA4+ Sertoli cells. At least 34 tubules were counted. (G) Ratio of KIT+ spermatogonia and GATA4+ Sertoli cells. At least 8 tubules were counted. (H) The number of TUNEL+ cells in seminiferous tubules. At least 11 tubules were counted. (I) The number of KIT+ cells undergoing apoptosis. At least 8 tubules were counted. (J) The number of EPCAM+ cells undergoing apoptosis. At least 10 tubules were counted. (K) Expression of MKI67 in CDH1+ cells. Eighteen tubules were counted. (L) Expression of MKI67 protein in KIT+ cells. Twenty tubules were counted. (Scale bar: B, 50 μm.) Stain, hematoxylin/eosin (B).
Double immunohistochemistry of the testes showed that the frequency of FBXW7-expressing cells in CDH1+ spermatogonia was reduced from 20.1% in the control to 3.2% in Fbxw7f/f Stra8-Cre mice, suggesting that the deletion efficiency was 84.1% (Fig. 3D and Fig. S4A). We also found that expression of synaptonemal complex protein 3 (SYCP3), a marker for meiotic germ cells, was significantly suppressed in these mice (Fig. 3E and Fig. S4B). On the other hand, the seminiferous tubules of Fbxw7f/f Stra8-Cre mice contained a high number of CDH1+ spermatogonia per Sertoli cells, as evaluated by the ratio of CDH1+ to GATA binding protein 4 (GATA4)+ Sertoli cells (Fig. 3F and Fig. S4C). Similar results are found for KIT (Fig. 3G and Fig. S4D), suggesting that FBXW7 plays a role in restricting the proliferation of spermatogonia. Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining showed increased apoptosis in EPCAM+ cells, but not in KIT+ cells (Fig. 3 H–J and Fig. S4 E and F). Because EPCAM is expressed on spermatogonia and KIT is expressed on germ cells up to the pachytene stage, this observation suggests that spermatogonia are predominantly undergoing apoptosis. We also observed a greater frequency of MKI67+ cells in both CDH1- and KIT-expressing spermatogonia in Fbxw7f/f Stra8-Cre mice (Fig. 3 K and L and Fig. S4 G and H). These results suggest that, in Fbxw7f/f Stra8-Cre mice, Fbxw7 deletion not only induces the proliferation of CDH1- and KIT-expressing spermatogonia but also causes increased apoptosis of premeiotic germ cells.
Enhanced SSC Activity of Fbxw7-Deficient Testis Cells.
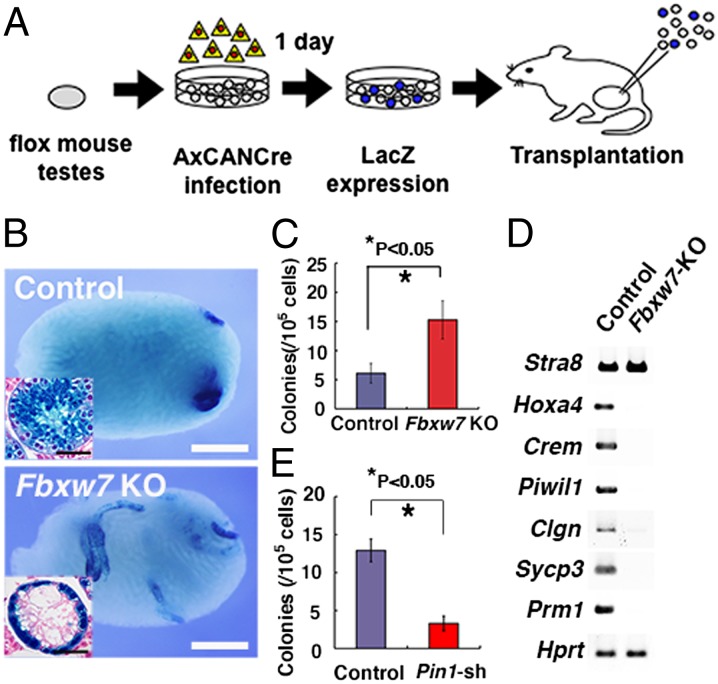
To examine whether Fbxw7 deficiency has any effect on SSCs, we performed transplantation experiments because SSCs cannot be identified by morphological analysis. Fbxw7 conditional knockout (KO) mice were crossed with a ROSA26 reporter mouse strain (R26R) to visualize the pattern of colonization (16). Testis cells were collected from 8- to 11-d-old Fbxw7f/f mice heterozygous for the R26R allele, and a single-cell suspension was exposed to a Cre-expressing adenovirus (AxCANCre) overnight in vitro and then injected into the seminiferous tubules of W mice (Fig. 4A and Fig. S5A).
Fig. 4.
Enhanced SSC activity in Fbxw7 KO testis cells. (A) Experimental procedure. Testis cells from Fbxw7f/f mice were dissociated and incubated with AxCANCre overnight and then injected into the seminiferous tubules of W testes. (B) Macroscopic appearance of recipient testis. Histological appearance (Inset). (C) Colony counts after Cre transfection and transplantation of Fbxw7f/f mouse testes. Results of three experiments (n = 18). (D) RT-PCR analysis of spermatogenic gene expression in recipient testes. (E) Colony counts after Pin1 depletion and transplantation of green mouse testes. Results of two experiments (n = 16). (Scale bars: B, 1 mm; B, Inset, 50 μm.)
After an overnight incubation, 87.7 ± 7.1% (n = 4) and 84.3 ± 6.9% (n = 6) of the infected cells were recovered from control and mutant testis cells, respectively, showing no significant difference. Southern blot analyses showed that 63.6 ± 2.0% (n = 4) of the floxed allele was deleted at the time of transplantation (Fig. S5B). Analysis of the recipient testes between 6 and 8 wk showed that the testis cells from Cre-treated Fbxw7f/f mice produced significantly more colonies than did control testis cells (6.1 ± 1.7 and 15.3 ± 3.2 colonies per 105 cells injected for control and Fbxw7f/f mice, respectively; n = 18) (Fig. 4 B and C). Histological analyses showed that all of the 282 colonized tubules counted contained only a single layer of LacZ+ cells on the basement membrane, and no apparent meiotic cells were found (Fig. 4B). Tumor formation was not observed in any of the recipient testes 6 mo after transplantation (n = 8). RT-PCR analysis of recipient testes showed that expression of meiotic germ cell markers, such as Hoxa4 and Sycp3, was significantly reduced or not detected (Fig. 4D).
Although Fbxw7 deficiency increased the concentration of SSCs, this result appeared to contradict a previous observation that Pin1 deficiency induced spermatogonia depletion (17), given that Pin1 depletion caused a decrease in FBXW7 level. Because PIN1 is also expressed in Sertoli cells and may influence spermatogenesis, we directly examined the impact of Pin1 depletion in germ cells by spermatogonial transplantation. We transduced testis cells from 9-d-old green mouse pups with shRNA against Pin1, and the cells were transplanted into W mice 2 d later. Analysis of the recipient mice showed significantly reduced colonies with Pin1 depletion (Fig. 4E). The number of colonies generated by cells transduced with control and Pin1 shRNA was 12.9 ± 1.5 and 3.3 ± 1.0 per 105 cells (n = 16). This result suggests that PIN1 expression in germ cells is essential for SSC activity and that PIN1 has additional targets involved in self-renewal.
Screening of Fbxw7 Target Proteins Using GS Cells.
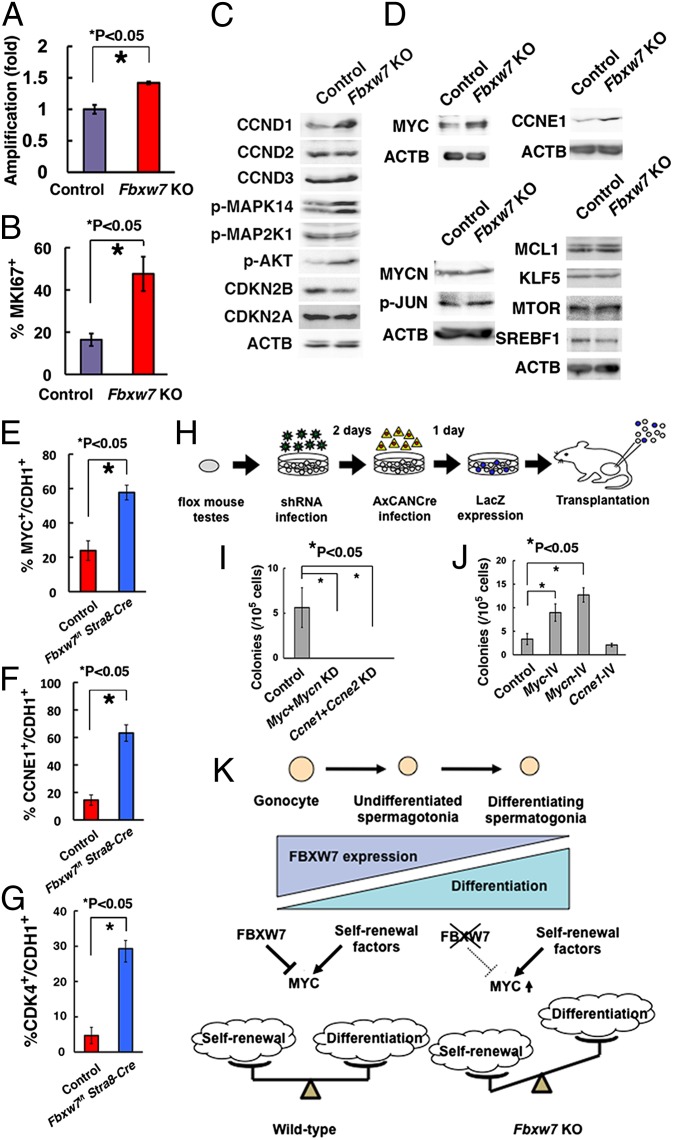
To investigate the molecular machinery involved in FBXW7-mediated self-renewal regulation, Fbxw7f/f GS cells were established and exposed in vitro to AxCANCre overnight. Unlike the deletion of primary testis cells, Southern blot analyses showed that the floxed allele was completely deleted from the Fbxw7 gene locus, which likely reflected enhanced proliferation of the Fbxw7 KO population (Fig. S6A). Consistent with this observation, the amplification rate was increased significantly after Cre-mediated deletion of Fbxw7 gene (Fig. 5A), which was supported by the increased frequency of MKI67+ cells in mutant cells (Fig. 5B and Fig. S6B). Flow cytometric analysis showed no significant changes in the expression of known spermatogonia markers (Fig. S6C).
Fig. 5.
Characterization of Fbxw7 KO GS cells for target identification. (A) Enhanced proliferation of GS cells after Cre transfection. After overnight incubation with AxCANCre, virus supernatant was removed, and cells were replated in a new dish. Cell number was determined 3 d after replating. AxCANLacZ was used as a control (n = 4). Results of two experiments. (B) Quantification of GS cells expressing MKI67. At least 732 cells in four different fields were counted. (C) Western blot analysis of molecules involved in proliferation of GS cells. (D) Western blot analysis of FBXW7 substrates. (E) Expression of MYC in CDH1+ cells in Fbxw7f/f Stra8-Cre testes. Fifteen tubules were counted. (F) Expression of CCNE1 in CDH1+ cells in Fbxw7f/f Stra8-Cre testes. Fifteen tubules were counted. (G) Expression of CDK4 in CDH1 cells in Fbxw7f/f Stra8-Cre testes. At least 8 tubules were counted. (H) Experimental procedure. Immature testis cells from Fbxw7f/f mice were infected with lentiviruses expressing shRNA against Myc/Mycn or Ccne1/Ccne2. Culture medium was changed on the next day after infection. Two days after shRNA infection, cells were infected with AxCANCre and transplanted 24 h after AxCANCre infection. (I) Colony counts after depleting the indicated genes (n = 8). Results of two experiments. (J) Colony counts after overexpression of indicated genes and transplantation of green mouse testis cells. Results of two experiments (n = 12). (K) Summary of experiments. Loss of FBXW7 increases self-renewal by up-regulation of MYC and suppresses differentiation.
To investigate molecules involved in enhanced spermatogonial proliferation, we first examined the expression of molecules involved in cell proliferation using Fbxw7 KO GS cells (Fig. S6D). We found not only increased expression of CCND1, phosphorylated MAPK14 (p-MAPK14), and AKT (p-AKT), but also decreased expression of cyclin-dependent kinase inhibitor 2B (CDKN2B) (Fig. 5C). We then analyzed the expression of candidate substrates of FBXW7. The expression of neither NOTCH1 nor NOTCH2 was enhanced by Fbxw7 deficiency (Fig. S6E). Levels of NICD (Notch intracellular domain) did not show significant changes (Fig. S6F). Moreover, adding a γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) or depletion of Rbpj, the common DNA binding partner of all Notch receptors, by shRNA did not influence proliferation of Fbxw7 KO GS cells (Fig. S6 G–I). We also checked several NOTCH target genes by real-time PCR. Although Hes1 expression was slightly increased, the rest of the genes did not change significantly by Fbxw7 deficiency (Fig. S6J). In contrast, expression of myelocytomatosis oncogene (MYC) and cyclin E1 (CCNE1) was enhanced in Fbxw7 KO GS cells (Fig. 5D and Fig. S6K). The expression levels of phosphorylated JUN (p-JUN), v-myc myelocytomatosis viral related oncogene, reuroblastoma derived (MYCN), SREBF1, MCL1, and KLF5 did not change significantly (Fig. 5D and Fig. S6K). We also did not detect changes in MTOR level, which reportedly influenced the expression of GDNF receptor components (18). These results suggest that MYC and CCNE1 are responsible for enhanced self-renewal of SSCs.
Myc Is Responsible for Enhanced SSC Activity by Fbxw7 Deficiency.
Consistent with these observations, double immunohistochemistry revealed significantly enhanced expression of MYC and CCNE1 in CDH1+ undifferentiated spermatogonia in Fbxw7f/f Stra8-Cre mice (Fig. 5 E and F and Fig. S7 A and B). When we examined several Myc classic target genes, we noted increased CDK4 expression in Fbxw7f/f Stra8-Cre mice (Fig. 5G and Fig. S7 C–E). To examine whether the accumulation of MYC and/or CCNE1 caused enhanced colonization after Fbxw7 deletion, we investigated the effect of Myc/Mycn knockdown (KD) using Fbxw7 KO mice. We used Mycn and Ccne2 shRNA because loss of Myc or Ccne1 may be compensated by these treatments (19, 20). Testis cells from 7-d-old Fbxw7f/f R26R mice were transduced with lentivirus expressing shRNA for Myc/Mycn or Ccne1/Ccne2 by overnight incubation, followed by treatment with AxCANCre 2 d after lentiviral infection. After overnight incubation with AxCANCre, cells were injected into W testes (Fig. 5H). Colonization was significantly suppressed by either Myc/Mycn or Ccne1/Ccne2 KD (Fig. 5I and Fig. S7F), suggesting positive roles for these molecules in SSC self-renewal.
Finally, we examined the impact of target gene overexpression. Testis cells from 8-d-old green mice were transduced with a lentivirus expressing Myc, Mycn, or Ccne1. After overnight infection, the viral supernatant was removed, and the cells were transplanted into seminiferous tubules of W mice the following day. Analysis of the recipient mice showed enhanced colonization of donor cells transduced with Myc or Mycn. In contrast, Ccne1 overexpression did not affect colonization (Fig. 5J and Fig. S7G). Myc overexpression could also increase colonization of pup testis transduced with Fbxw7 (Fig. S7H). Although Pin1 depletion did not change MYC in GS cells, it significantly enhanced CCNE1 (Fig. S7 I and J). Myc silencing in Pin1-depleted cells did not induce significant changes in SSC activity (Fig. S7K). Because Pin1 depletion decreased SSC activity (Fig. 4E), these results suggest that MYC mediates the enhanced SSC activity seen with Fbxw7 deficiency.
Discussion
Recent studies revealed that Fbxw7 plays pivotal roles in the regulation of several stem cells/progenitors (21–25). Most of the phenotypes of Fbxw7 deficiency are related to cell proliferation, death, or skewed differentiation. However, effects on stem cell self-renewal are not always evident and can be variable. In testes, FBXW7 expression is restricted to a subset of spermatogonia in a cell cycle-specific manner. Fbxw7 mRNA expression was down-regulated by Zbtb16 depletion whereas it was up-regulated by Id2/Id3/Id4 depletion. Increased Fbxw7 expression by Id2/Id3/Id4 depletion was intriguing. Given that ID proteins often influence cell-cycle progression (26), it is possible that ID proteins may play a role in regulating FBXW7 expression, which occurred in a cell cycle-dependent manner. However, depletion of these molecules did not lead to apparent changes in Western blots. Our screening further revealed that FBXW7 down-regulation occurs upon depletion of Pin1, whose deficiency causes male infertility (17). This result was unexpected because PIN1 is thought to promote FBXW7 degradation by causing self-ubiquitination (14). Although Pin1 overexpression did not change FBXW7 expression, it is possible that the amount of PIN1 overexpression was not sufficient to influence FBXW7. Alternatively, PIN1 may collaborate with other molecules. Because promotion of FBXW7 degradation by PIN1 was demonstrated in cancer cells, the interaction between FBXW7 and PIN1 may be more complex in nontransformed spermatogonia and may involve additional molecules. Fbxw7 is a tumor suppressor, and its expression needs to be tightly controlled in nontransformed cells. Indeed, PIN1 expression is inversely correlated with FBXW7 expression (14) whereas PIN1 expression was enhanced in undifferentiated relative to differentiating spermatogonia (17), suggesting another sophisticated regulation system. Because Pin1 depletion compromised SSC activity, we speculate that the balance between PIN1 and FBXW7 is critical in determining SSC fate and that disruption of this regulation may induce tumor formation. Our results suggest that PIN1 functions in a context-dependent manner to regulate FBXW7 expression.
We found that overexpression of Fbxw7α compromised SSC activity of testis cells. Although SSC number increased dramatically in pup testes (27), donor-cell colonization was significantly decreased by Fbxw7α overexpression. Given the strong growth suppression of GS cells by Fbxw7α overexpression and its cell cycle-specific expression, this result suggests that the amount and timing of FBXW7 need to be tightly controlled to promote cell-cycle progression. It also suggests that committed progenitors proliferate faster than undifferentiated spermatogonia because they lack FBXW7 expression. However, we currently cannot explain why Fbxw7α overexpression reduced the colonization activity of SSCs. It might be related to the fact that FBXW7 is restricted to undifferentiated spermatogonia in the spermatogenic system. One possibility is that Fbxw7α -overexpressing SSCs may have seeded in the recipient testes but ectopic Fbxw7α overexpression in progenitors may have hindered colony formation. It is also possible that SSCs may have failed to colonize the recipient tubules due to cell-cycle dysregulation, which is one of the important factors involved in migration of SSCs to niches (28).
Our results showed that Fbxw7 deletion increased proliferation of undifferentiated spermatogonia. Importantly, transplantation experiments confirmed its effect on SSCs. Together with the results from Fbxw7 overexpression experiments, these results demonstrate that FBXW7 plays an important role in negatively regulating self-renewal. To our knowledge, this is the first report of increased SSC activity in KO mice. Although enhanced spermatogonial proliferation was observed in Cdkn1b KO mice (29), Cdkn1b is different from Fbxw7 in that its deficiency promoted differentiating division and a decreased rather than increased SSC frequency. Tsc22d3 deficiency was also associated with increased spermatogonia proliferation, but undifferentiated spermatogonia were lost in the adult (30). Mutations in other genes, such as Sohlh1/Sohlh2, suppressed differentiation (31), but Fbxw7 is unique because its deficiency not only caused enhanced proliferation of undifferentiated spermatogonia but also increased self-renewal division. In this regard, it should be noted here that enhancement of self-renewal activity was accompanied by impaired differentiation and increased apoptosis. Although Fbxw7 deficiency often caused apoptosis and skewed differentiation, complete suppression of differentiation has not been observed in other self-renewing tissues. In this sense, the spermatogenic system appears to be extremely sensitive to the amount of FBXW7 in differentiation.
In our search for candidate FBXW7 target molecules, we found up-regulation of MYC and CCNE1 in Fbxw7 KO GS cells. Myc plays a key role in exit from and reentry into the cell cycle, and its overexpression was also implicated in human tumorigenesis (32). The function of Mycn in spermatogonia was reported in a previous study (33), which showed that Mycn is up-regulated by GDNF in a phosphatidylinositol 3-kinase-AKT dependent manner to contribute to proliferation. However, because this study was carried out in an SV40-transformed spermatogonia cell line, the role of Myc family genes in SSCs has remained unclear. The importance of Ccne1 in spermatogonia was demonstrated in several studies. Ccne1 overexpression increased SSC self-renewal by cooperating with Ccnd2 (7) whereas Ccne2 deficiency compromised spermatogenesis (19).
Several lines of evidence suggest that MYC is a target substrate of FBXW7 in SSCs and that it increases SSC activity. First, loss of Fbxw7 induced accumulation of MYC in undifferentiated spermatogonia and GS cells. Second, suppression of Myc/Mycn by shRNAs abolished the up-regulated SSC activity caused by Fbxw7 deficiency. Third, Myc or Mycn overexpression increased the colonization efficiency of wild-type (WT) testis cells. Although depletion of Ccne1 and Ccne2 expression could reduce SSC activity, Ccne1 overexpression did not enhance colonization, suggesting that CCNE1/CCNE2 are not involved in the abnormal phenotype of Fbxw7 KO mice. Enhanced apoptosis in Fbxw7-deficient testis may also be caused by increased MYC expression (32). Taken together, these lines of evidence strongly suggest that FBXW7 counteracts with positive regulators of self-renewal by degrading MYC (Fig. 5K).
Although the current study identified a role for FBXW7 as a negative regulator of SSC self-renewal, several questions persist. For example, although PIN1 is thought to bind to FBXW7 in a phosphorylation-dependent manner, we need to find targets for PIN1 and whether they have any interaction with FBXW7. We also do not know why FBXW7 influenced the expression of MYC, but not MYCN, and whether MYC up-regulation is involved in the suppression of spermatogonial differentiation. Having identified, to our knowledge, the first negative regulator of SSCs, we are now at the stage to find other negative regulators that may counteract with self-renewal signals at different levels. In addition, modulation of FBXW7 function by small molecules may be useful for enhancement of SSC self-renewal in vitro for genetic modifications. Thus, identification of the negative regulator for SSC self-renewal not only deepens our knowledge of SSC biology but also adds a new dimension of investigation and possibilities.
Materials and Methods
Animals.
The generation of Fbxw7f/f mice was described previously (34). WT C57BL/6 mice were purchased from Japan SLC. We also used 8- to 10-d-old green mice that ubiquitously express EGFP (a gift from D. M. Okabe, Osaka University, Osaka). Male Fbxw7f/f mice were crossed with R26R female mice (16) to introduce the LacZ reporter construct for Cre-mediated deletion (The Jackson Laboratory). Stra8-Cre transgenic mice were also purchased from The Jackson Laboratory. Genotypes of the mice were examined by PCR with the primers listed in Table S3.
Transplantation.
For transplantation, testis cells were dissociated into single-cell suspensions by a two-step enzymatic digestion using collagenase type IV and trypsin (Sigma), as described previously (35). GS cells were incubated with 0.25% trypsin to obtain single-cell suspensions. The donor cells were transplanted into seminiferous tubules of W mice (Japan SLC) through the efferent duct. Approximately 4 μL could be introduced into each testis, which filled 75–85% of the seminiferous tubules.
The Institutional Animal Care and Use Committee of Kyoto University approved all animal experimentation protocols. Further details of procedures are described in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ms. Y. Ogata for technical assistance. This research was supported by the government of Japan through its “Funding Program for Next Generation World-Leading Researchers,” the Japan Science and Technology Agency (Core Research for Evolutionary Science and Technology, Precursory Research for Embryonic Science and Technology), the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Takeda Science Foundation, the Uehara Memorial Foundation, and the Ministry of Education, Culture, Sports, Science, and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401837111/-/DCSupplemental.
References
- 1.de Rooij DG, Russell LD. All you wanted to know about spermatogonia but were afraid to ask. J Androl. 2000;21(6):776–798. [PubMed] [Google Scholar]
- 2.Meistrich ML, van Beek MEAB. Cell and molecular biology of the testis. In: Desjardins C, Ewing LL, editors. Spermatogonial Stem Cells. New York: Oxford Univ Press; 1993. pp. 266–295. [Google Scholar]
- 3.Meng X, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287(5457):1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 4.Tegelenbosch RAJ, de Rooij DG. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat Res. 1993;290(2):193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- 5.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA. 1994;91(24):11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanatsu-Shinohara M, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69(2):612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 7.Lee J, et al. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell. 2009;5(1):76–86. doi: 10.1016/j.stem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Ishii K, Kanatsu-Shinohara M, Toyokuni S, Shinohara T. FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development. 2012;139(10):1734–1743. doi: 10.1242/dev.076539. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, et al. Akt mediates self-renewal division of mouse spermatogonial stem cells. Development. 2007;134(10):1853–1859. doi: 10.1242/dev.003004. [DOI] [PubMed] [Google Scholar]
- 10.Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem. 2007;282(35):25842–25851. doi: 10.1074/jbc.M703474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA. 2006;103(25):9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welcker M, Clurman BE. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 13.Pashkova N, et al. WD40 repeat propellers define a ubiquitin-binding domain that regulates turnover of F box proteins. Mol Cell. 2010;40(3):433–443. doi: 10.1016/j.molcel.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min S-H, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46(6):771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46(12):738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 17.Atchison FW, Means AR. Spermatogonial depletion in adult Pin1-deficient mice. Biol Reprod. 2003;69(6):1989–1997. doi: 10.1095/biolreprod.103.020859. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142(3):468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114(4):431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 20.Laurenti E, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3(6):611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babaei-Jadidi R, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208(2):295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoeck JD, et al. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci. 2010;13(11):1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- 23.Matsumoto A, et al. Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J Biol Chem. 2011;286(15):13754–13764. doi: 10.1074/jbc.M110.194936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka S, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22(8):986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancho R, et al. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology. 2010;139(3):929–941. doi: 10.1053/j.gastro.2010.05.078. [DOI] [PubMed] [Google Scholar]
- 26.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13(8):410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 27.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Remodeling of the postnatal mouse testis is accompanied by dramatic changes in stem cell number and niche accessibility. Proc Natl Acad Sci USA. 2001;98(11):6186–6191. doi: 10.1073/pnas.111158198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishii K, Kanatsu-Shinohara M, Shinohara T. Cell-cycle-dependent colonization of mouse spermatogonial stem cells after transplantation into seminiferous tubules. J Reprod Dev. 2014;60(1):37–46. doi: 10.1262/jrd.2013-083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanatsu-Shinohara M, Takashima S, Shinohara T. Transmission distortion by loss of p21 or p27 cyclin-dependent kinase inhibitors following competitive spermatogonial transplantation. Proc Natl Acad Sci USA. 2010;107(14):6210–6215. doi: 10.1073/pnas.0914448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruscoli S, et al. Long glucocorticoid-induced leucine zipper (L-GILZ) protein interacts with ras protein pathway and contributes to spermatogenesis control. J Biol Chem. 2012;287(2):1242–1251. doi: 10.1074/jbc.M111.316372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki H, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361(2):301–312. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braydich-Stolle L, Kostereva N, Dym M, Hofmann M-C. Role of Src family kinases and N-Myc in spermatogonial stem cell proliferation. Dev Biol. 2007;304(1):34–45. doi: 10.1016/j.ydbio.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onoyama I, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204(12):2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int J Dev Biol. 1997;41(1):111–122. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.