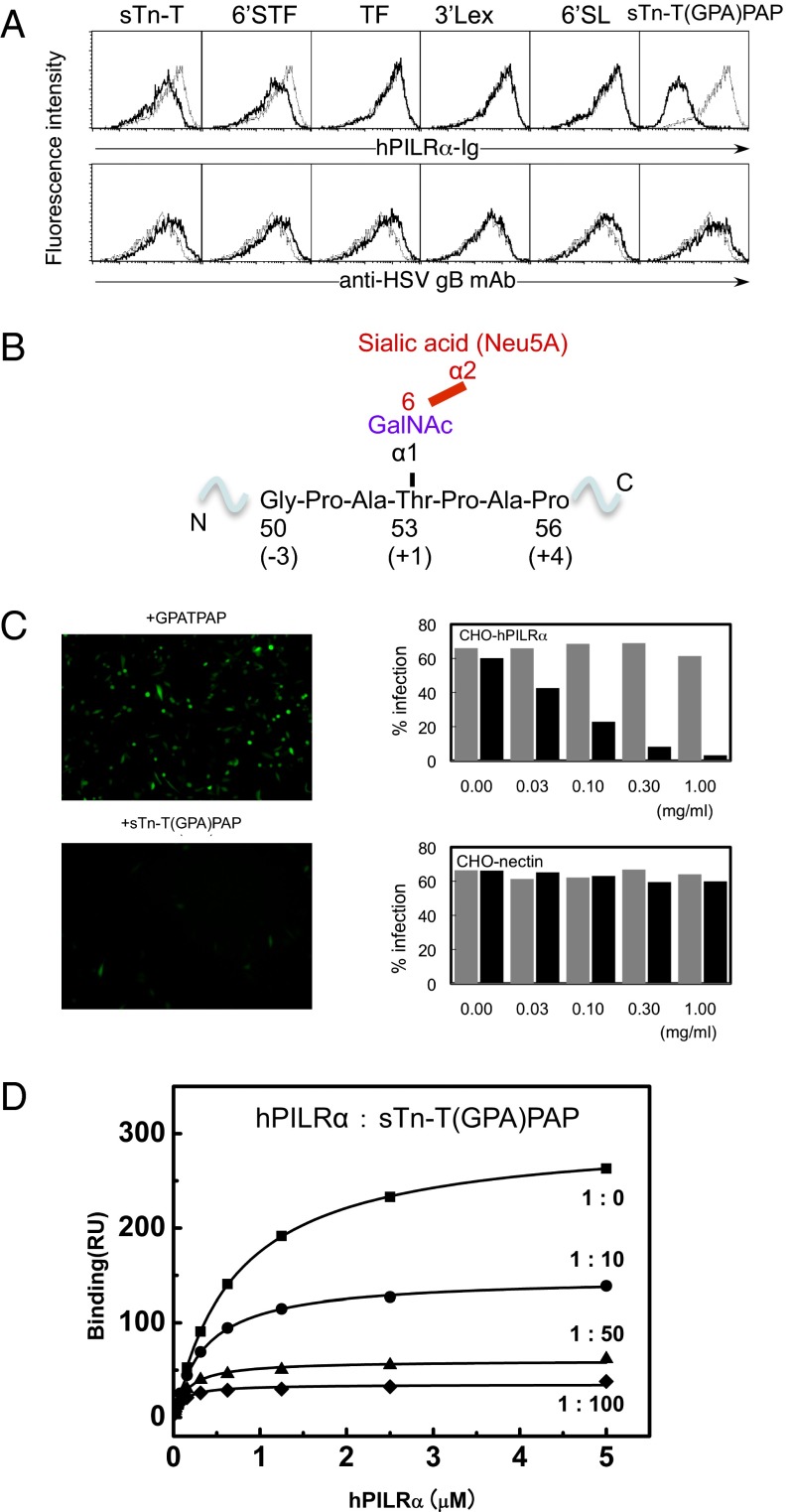
Fig. 1.
Identification of the region in HSV-1 gB that is required for PILRα recognition. (A) Competition assay with various sugar compounds assessing the ability of the PILRα-Ig fusion protein to bind to HEK293 cells expressing the HSV-1 gB molecule. The black and gray lines indicate binding in the presence or absence of additional sugar compounds (final concentration, 0.5 mg/mL), respectively. sTn-T, Neu5Acα2–6GalNAcα-O-Thr; 6′STF, Galβ1–3[Neu5Acα2–6]GalNAcα-O-Thr; TF, Galβ1–3GalNAcα-O-Thr; 3′SLex, Neu5Acα2–6Galβ1–4[Fucα1–3]GalNAcβ-Sp; 6′SL, Neu5Acα2–6Galβ1–4Glcβ-Sp. (B) The detailed structure surrounding Thr53 of gB with an O-linked sugar and the region that represents the sTn-T(GPA)PAP peptide. (C) Effects of sTn-T(GPA)PAP or GPATPAP on HSV-1 infection. (Left) Representative images. (Right) Dose-dependent effects of sTn-T(GPA)PAP (black) or GPATPAP (gray) on the infection efficiency in CHO cells expressing PILRα or nectin-1. (D) SPR competition assay examining the ability of the sTn-T(GPA)PAP glycopeptide to inhibit PILRα binding to HSV-1 gB. Closed squares, PILRα only; closed circles, PILRα:sTn peptide, 1:10; closed triangles, PILRα:sTn peptide, 1:50; closed diamonds, PILRα:sTn peptide, 1:100.