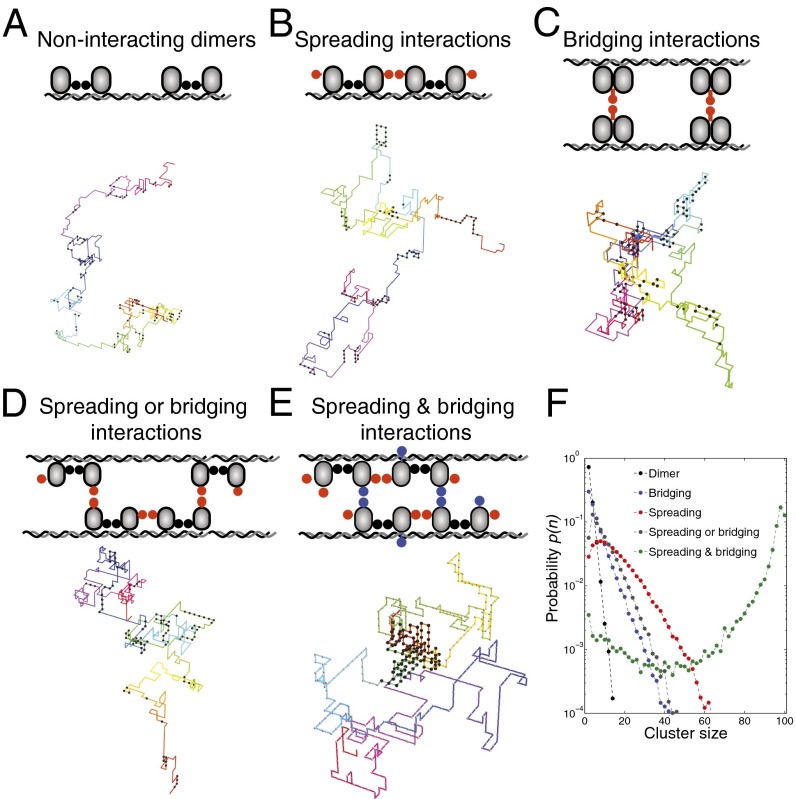
Fig. 2.
Simple models for interacting DNA-bound proteins. Schematics include a representative image of a DNA–protein complex from simulations: (A) dimer model, (B) spreading model, (C) bridging model, (D) spreading or bridging model, and (E) spreading and bridging model. In the spreading or bridging model there are two interaction domains per monomer, one of which can be either in spreading or in bridging mode. In the spreading and bridging model there are two 1D spreading bonds along the DNA and one 3D bridging bond. The bond energies are except in the spreading or bridging model, for which the bridging bond has an energy of . The bending rigidity of the DNA was set to . The total number of proteins is fixed at on a DNA chain of length (i.e., with 500 binding sites). (F) Probability distribution for a DNA-bound protein to be part of a cluster of size n.