Fig. 5.
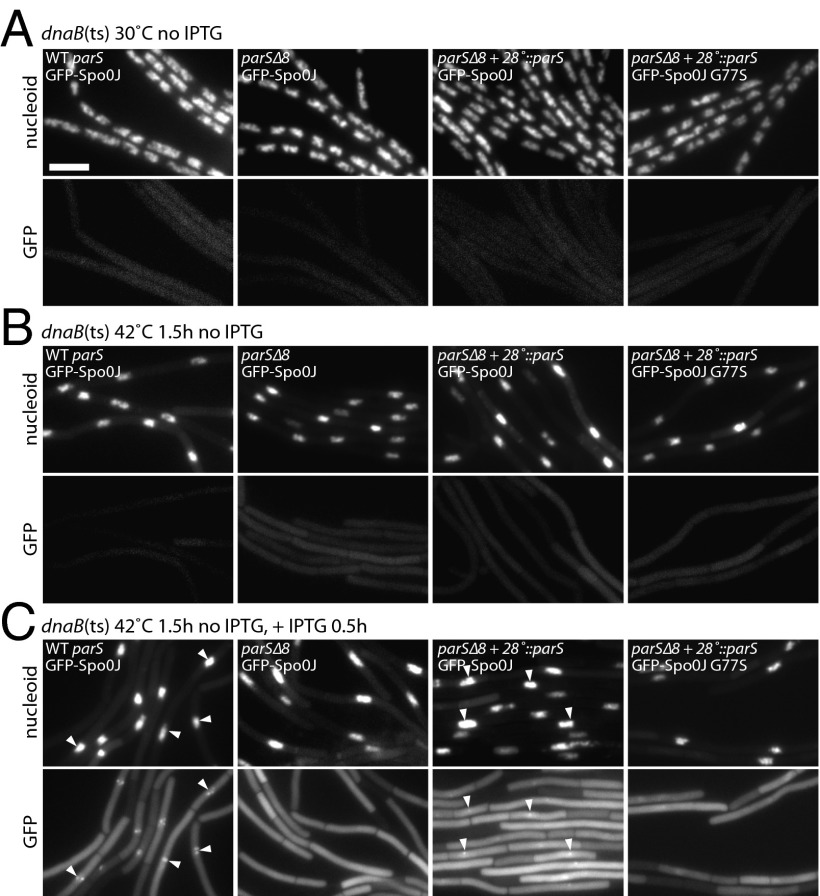
A single parS site is necessary and sufficient to generate a GFP-Spo0J focus. (A) Representative images of B. subtilis cells harboring a dnaB(ts) allele with all eight wild-type parS sites (BWX2454, WT parS, first panel); none of the eight endogenous parS sites (BWX2456, parSΔ8, second panel); parSΔ8 with an ectopic parS site inserted at 28° (amyE) (BWX2458 and BWX2789, parSΔ8+28°::parS, third and fourth panels) grown at 30 °C (A) and after growth at 42 °C for 1.5 h (B) to block new rounds of initiation of replication generating a single chromosome (Supporting Information). Expression of GFP-Spo0J (first to third panels) or GFP-Spo0J (G77S) (fourth panel) was then induced by the addition of IPTG (0.5 mM final concentration) for 0.5 h at 42 °C (C) (see Supporting Information for induction control). GFP-Spo0J did not form foci in the strain lacking parS sites (parSΔ8, second panel) and formed a single focus per nucleoid in the parSΔ8+28°::parS strain (third panel). GFP-Spo0J (G77S) did not form foci in theparSΔ8+28°::parS strain (fourth panel). Based on comparison with Spo0J (G77S) (C, fourth panel) that binds parS but is unable to spread to neighboring sites (13), the foci observed in the parSΔ8+28°::parS strain (C, third panel) reflect GFP-Spo0J nucleoprotein complexes. Gray scale of DAPI-stained nucleoids (upper panels) and GFP (lower panels) is shown. GFP-Spo0J foci (white carets) are highlighted in the DAPI and GFP panels. (Scale bar, 4 μm.)