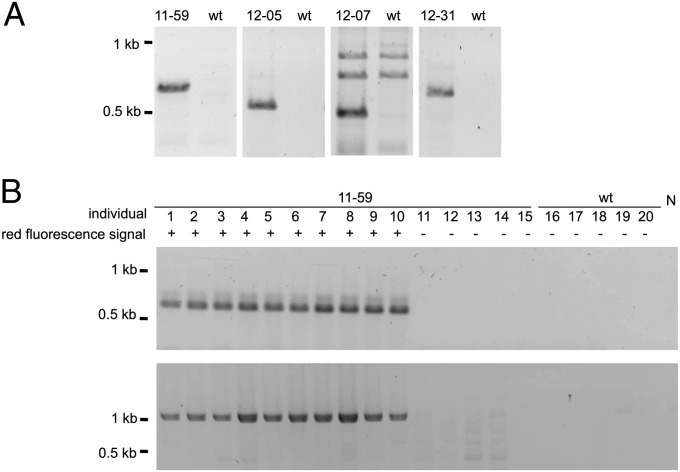
Fig. 2.
Stable genomic integration and transmission of the [6xP3-rubia] expression cassette. (A) The different genomic integration sites of the [6xP3-rubia] expression cassette in the four queens as revealed by genomic loci-specific PCRs. The different genomic integration sites were amplified from DNA using a genome- and a cassette-specific oligonucleotide primer in the PCRs. The fragments were separated by agarose gel electrophoresis. The specific oligonucleotide primer pairs used were as follows for the four queens: queen 11–59: #178/#090 (expected size: 586 bp); queen 12–05: #206/#090 (expected size: 428 bp); queen 12–07: #201/#090 (expected size: 529 bp); and queen 12–31: #204/#090 (expected size: 453 bp). The DNA template, representing the queen’s genotype, was obtained by pooling the haploid drone offspring. wt denotes a pool of wild-type honeybee drones. (B) Example of stable integration and transmission for the [6xP3-rubia] cassette in single offspring of queen 11–59. We amplified the sequence from DNA of single offspring in two separate PCRs; one amplifying specifically the integration site (Upper gel image) and one amplifying only the expression cassette alone (Lower gel image). Fragments were separated by agarose gel electrophoresis. Offspring of queen 11–59 that exhibited a red fluorescent signal of the transgenic marker (as indicated by +) had the expression cassette integrated at the same predicted genomic integration site. Offspring that exhibited no red fluorescent signal of the transgenic marker (as indicated by –) possessed no [6xP3-rubia] expression cassette. N denotes the control reaction with no DNA. The gels were stained with ethidium bromide. The identities of the fragments were verified by sequencing.